Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1227
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3327 | 1.554e-03 | 2.800e-05 | 1.645e-01 | 7.157e-09 |
|---|
| IPC | 0.3386 | 3.524e-01 | 1.816e-01 | 9.364e-01 | 5.995e-02 |
|---|
| Loi | 0.4851 | 3.600e-05 | 0.000e+00 | 4.914e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6002 | 1.000e-06 | 0.000e+00 | 1.123e-01 | 0.000e+00 |
|---|
| Wang | 0.2568 | 7.787e-03 | 6.709e-02 | 2.319e-01 | 1.211e-04 |
|---|
Expression data for subnetwork 1227 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
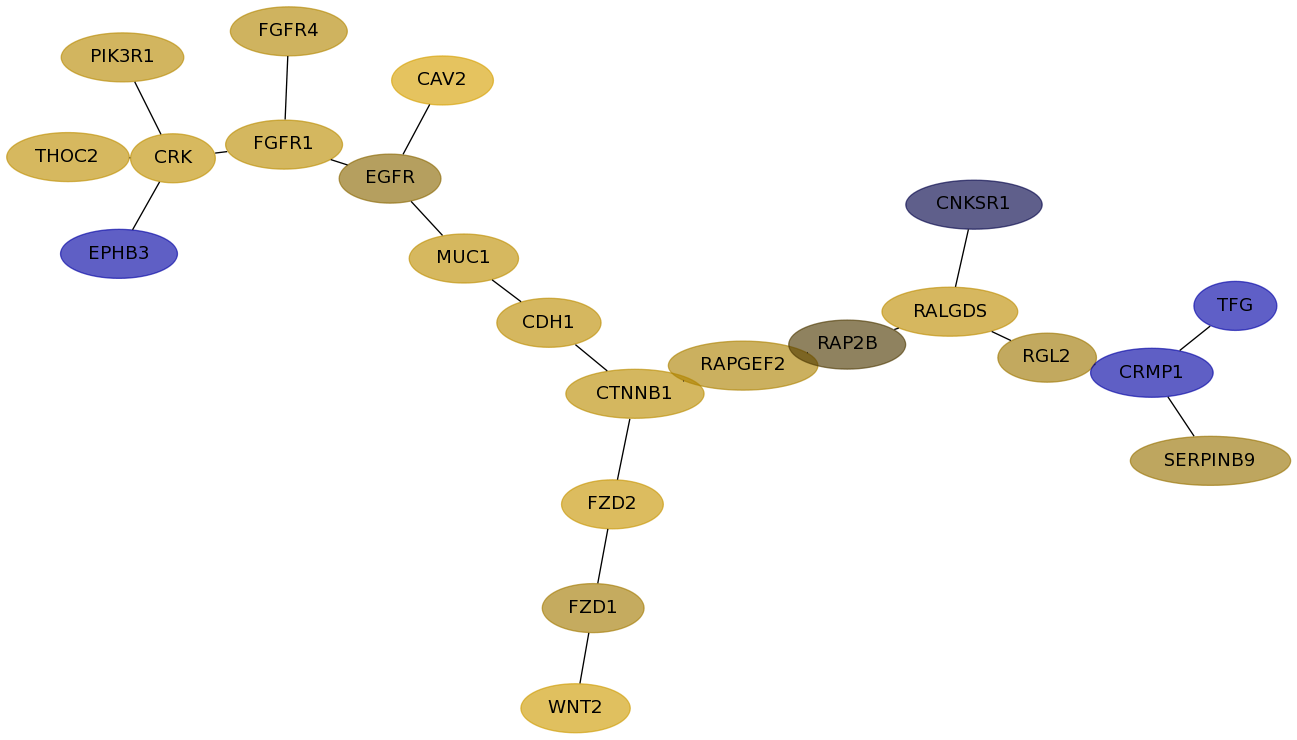
Score for each gene in subnetwork 1227 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| RAPGEF2 |   | 1 | 652 | 647 | 664 | 0.144 | -0.245 | 0.202 | -0.019 | 0.210 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| WNT2 |   | 2 | 391 | 647 | 646 | 0.314 | 0.223 | -0.064 | -0.148 | 0.186 |
|---|
| SERPINB9 |   | 2 | 391 | 647 | 646 | 0.091 | -0.094 | -0.073 | -0.025 | 0.002 |
|---|
| RAP2B |   | 1 | 652 | 647 | 664 | 0.015 | 0.118 | -0.040 | 0.004 | 0.062 |
|---|
| RALGDS |   | 5 | 148 | 647 | 621 | 0.212 | -0.011 | 0.199 | 0.033 | -0.009 |
|---|
| CNKSR1 |   | 2 | 391 | 647 | 646 | -0.013 | -0.025 | 0.209 | -0.012 | 0.036 |
|---|
| FZD2 |   | 8 | 84 | 552 | 540 | 0.273 | -0.067 | -0.081 | -0.067 | 0.081 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| THOC2 |   | 11 | 59 | 419 | 406 | 0.217 | -0.094 | 0.119 | 0.068 | 0.072 |
|---|
| RGL2 |   | 3 | 265 | 647 | 632 | 0.105 | -0.107 | 0.227 | -0.146 | -0.002 |
|---|
| TFG |   | 1 | 652 | 647 | 664 | -0.124 | -0.040 | 0.155 | -0.191 | 0.238 |
|---|
| CRMP1 |   | 3 | 265 | 647 | 632 | -0.114 | -0.068 | 0.174 | 0.049 | 0.151 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| FZD1 |   | 2 | 391 | 647 | 646 | 0.117 | -0.281 | 0.189 | -0.114 | -0.023 |
|---|
| EPHB3 |   | 4 | 198 | 326 | 333 | -0.115 | -0.048 | 0.135 | 0.099 | -0.008 |
|---|
GO Enrichment output for subnetwork 1227 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.699E-07 | 3.907E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 3.391E-07 | 3.9E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 3.391E-07 | 2.6E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.416E-06 | 8.144E-04 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 3.172E-06 | 1.459E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.787E-06 | 1.835E-03 |
|---|
| negative regulation of endothelial cell proliferation | GO:0001937 |  | 6.081E-06 | 1.998E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.353E-05 | 3.891E-03 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 1.604E-05 | 4.099E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.884E-05 | 4.332E-03 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 3.32E-05 | 6.941E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.848E-07 | 4.514E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.953E-07 | 3.607E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.953E-07 | 2.405E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.667E-07 | 5.294E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.905E-06 | 9.307E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 2.378E-06 | 9.682E-04 |
|---|
| negative regulation of endothelial cell proliferation | GO:0001937 |  | 3.545E-06 | 1.237E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.249E-06 | 1.297E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.898E-06 | 1.873E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.046E-05 | 2.555E-03 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 1.046E-05 | 2.323E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.497E-07 | 3.602E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.616E-07 | 3.147E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.616E-07 | 2.098E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.931E-07 | 5.372E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 2.12E-06 | 1.02E-03 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 3.362E-06 | 1.348E-03 |
|---|
| negative regulation of endothelial cell proliferation | GO:0001937 |  | 4.132E-06 | 1.42E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 5.011E-06 | 1.507E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.365E-06 | 2.236E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.127E-05 | 2.711E-03 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 1.127E-05 | 2.465E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.699E-07 | 3.907E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 3.391E-07 | 3.9E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 3.391E-07 | 2.6E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.416E-06 | 8.144E-04 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 3.172E-06 | 1.459E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.787E-06 | 1.835E-03 |
|---|
| negative regulation of endothelial cell proliferation | GO:0001937 |  | 6.081E-06 | 1.998E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.353E-05 | 3.891E-03 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 1.604E-05 | 4.099E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.884E-05 | 4.332E-03 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 3.32E-05 | 6.941E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.699E-07 | 3.907E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 3.391E-07 | 3.9E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 3.391E-07 | 2.6E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.416E-06 | 8.144E-04 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 3.172E-06 | 1.459E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.787E-06 | 1.835E-03 |
|---|
| negative regulation of endothelial cell proliferation | GO:0001937 |  | 6.081E-06 | 1.998E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.353E-05 | 3.891E-03 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 1.604E-05 | 4.099E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.884E-05 | 4.332E-03 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 3.32E-05 | 6.941E-03 |
|---|


