Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1211
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3324 | 1.576e-03 | 2.800e-05 | 1.655e-01 | 7.301e-09 |
|---|
| IPC | 0.3417 | 3.473e-01 | 1.771e-01 | 9.353e-01 | 5.755e-02 |
|---|
| Loi | 0.4828 | 4.100e-05 | 0.000e+00 | 5.024e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6021 | 1.000e-06 | 0.000e+00 | 1.086e-01 | 0.000e+00 |
|---|
| Wang | 0.2574 | 7.614e-03 | 6.615e-02 | 2.295e-01 | 1.156e-04 |
|---|
Expression data for subnetwork 1211 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
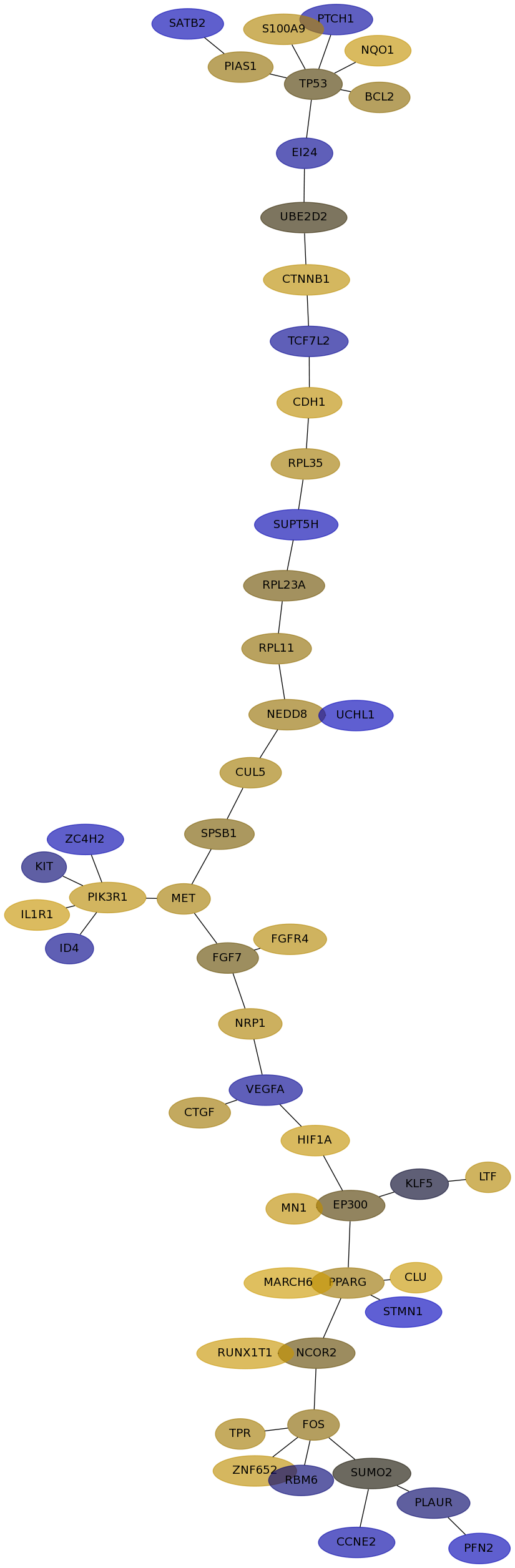
Score for each gene in subnetwork 1211 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| CCNE2 |   | 8 | 84 | 536 | 515 | -0.123 | 0.136 | -0.178 | 0.241 | -0.055 |
|---|
| NCOR2 |   | 11 | 59 | 283 | 274 | 0.024 | -0.096 | 0.211 | -0.163 | 0.114 |
|---|
| NQO1 |   | 6 | 120 | 536 | 535 | 0.243 | -0.115 | 0.069 | -0.189 | 0.118 |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| PFN2 |   | 4 | 198 | 182 | 183 | -0.164 | 0.014 | -0.001 | 0.286 | -0.056 |
|---|
| STMN1 |   | 1 | 652 | 536 | 560 | -0.199 | 0.168 | -0.033 | 0.198 | 0.062 |
|---|
| SPSB1 |   | 7 | 100 | 492 | 495 | 0.044 | -0.124 | 0.088 | -0.152 | 0.169 |
|---|
| LTF |   | 7 | 100 | 492 | 495 | 0.168 | -0.082 | 0.060 | -0.268 | -0.280 |
|---|
| RPL11 |   | 1 | 652 | 536 | 560 | 0.072 | -0.176 | 0.076 | 0.295 | -0.047 |
|---|
| RPL23A |   | 10 | 67 | 57 | 52 | 0.031 | 0.208 | -0.007 | 0.024 | 0.055 |
|---|
| PIAS1 |   | 1 | 652 | 536 | 560 | 0.076 | -0.270 | 0.096 | 0.157 | -0.083 |
|---|
| CUL5 |   | 4 | 198 | 492 | 499 | 0.111 | 0.051 | 0.187 | 0.221 | 0.216 |
|---|
| CTGF |   | 10 | 67 | 57 | 52 | 0.106 | -0.169 | 0.181 | 0.126 | 0.209 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| RBM6 |   | 10 | 67 | 148 | 152 | -0.036 | -0.082 | 0.265 | -0.004 | 0.111 |
|---|
| EI24 |   | 8 | 84 | 339 | 336 | -0.074 | 0.051 | 0.078 | 0.107 | 0.070 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| MET |   | 5 | 148 | 536 | 536 | 0.119 | -0.219 | 0.086 | 0.183 | 0.170 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| UCHL1 |   | 1 | 652 | 536 | 560 | -0.177 | -0.031 | -0.164 | 0.135 | 0.024 |
|---|
| MN1 |   | 6 | 120 | 148 | 156 | 0.213 | -0.166 | -0.047 | 0.150 | 0.011 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| RPL35 |   | 5 | 148 | 57 | 63 | 0.114 | 0.108 | -0.012 | 0.370 | -0.220 |
|---|
| RUNX1T1 |   | 3 | 265 | 536 | 537 | 0.271 | -0.170 | -0.040 | 0.081 | -0.099 |
|---|
| PPARG |   | 3 | 265 | 283 | 292 | 0.092 | -0.119 | 0.011 | 0.105 | 0.039 |
|---|
| PLAUR |   | 1 | 652 | 536 | 560 | -0.028 | 0.232 | 0.083 | 0.022 | 0.108 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PTCH1 |   | 4 | 198 | 339 | 341 | -0.102 | -0.184 | 0.047 | 0.121 | 0.004 |
|---|
| SATB2 |   | 1 | 652 | 536 | 560 | -0.157 | 0.222 | 0.233 | 0.166 | 0.050 |
|---|
| MARCH6 |   | 1 | 652 | 536 | 560 | 0.306 | -0.271 | 0.105 | 0.025 | -0.056 |
|---|
| SUPT5H |   | 9 | 77 | 1 | 18 | -0.150 | -0.085 | 0.007 | -0.298 | -0.068 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| IL1R1 |   | 8 | 84 | 536 | 515 | 0.257 | -0.211 | 0.178 | -0.071 | 0.003 |
|---|
| NEDD8 |   | 3 | 265 | 536 | 537 | 0.083 | 0.250 | -0.085 | 0.251 | -0.073 |
|---|
| TPR |   | 10 | 67 | 148 | 152 | 0.118 | -0.130 | 0.120 | -0.036 | 0.156 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| UBE2D2 |   | 2 | 391 | 536 | 545 | 0.007 | 0.212 | 0.002 | 0.217 | 0.090 |
|---|
| FGF7 |   | 8 | 84 | 398 | 384 | 0.026 | -0.094 | 0.237 | 0.070 | 0.083 |
|---|
| ZNF652 |   | 3 | 265 | 536 | 537 | 0.208 | -0.215 | -0.045 | -0.150 | -0.076 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| CLU |   | 10 | 67 | 117 | 121 | 0.280 | -0.208 | 0.119 | -0.153 | -0.234 |
|---|
| NRP1 |   | 2 | 391 | 536 | 545 | 0.152 | -0.043 | 0.025 | 0.201 | 0.071 |
|---|
GO Enrichment output for subnetwork 1211 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.694E-06 | 3.897E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 3.375E-06 | 3.881E-03 |
|---|
| negative regulation of growth | GO:0045926 |  | 3.918E-06 | 3.004E-03 |
|---|
| B cell lineage commitment | GO:0002326 |  | 5.882E-06 | 3.382E-03 |
|---|
| T cell lineage commitment | GO:0002360 |  | 5.882E-06 | 2.706E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.052E-06 | 3.47E-03 |
|---|
| negative regulation of cell development | GO:0010721 |  | 9.052E-06 | 2.974E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 9.052E-06 | 2.602E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.373E-06 | 2.395E-03 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 9.373E-06 | 2.156E-03 |
|---|
| cell fate commitment | GO:0045165 |  | 1.285E-05 | 2.687E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.451E-10 | 1.087E-06 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.962E-08 | 1.095E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.079E-07 | 8.79E-05 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 1.213E-07 | 7.406E-05 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 3.312E-07 | 1.618E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 3.861E-07 | 1.572E-04 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 5.067E-07 | 1.768E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 6.339E-07 | 1.936E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 6.552E-07 | 1.779E-04 |
|---|
| pancreas development | GO:0031016 |  | 1.157E-06 | 2.828E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.264E-06 | 2.807E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.052E-10 | 7.343E-07 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.231E-08 | 1.11E-04 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 1.454E-07 | 1.166E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 3.971E-07 | 2.388E-04 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 5.53E-07 | 2.661E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 7.201E-07 | 2.888E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 7.278E-07 | 2.502E-04 |
|---|
| pancreas development | GO:0031016 |  | 1.386E-06 | 4.17E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.451E-06 | 3.879E-04 |
|---|
| negative regulation of growth | GO:0045926 |  | 1.551E-06 | 3.731E-04 |
|---|
| B cell lineage commitment | GO:0002326 |  | 2.532E-06 | 5.537E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.694E-06 | 3.897E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 3.375E-06 | 3.881E-03 |
|---|
| negative regulation of growth | GO:0045926 |  | 3.918E-06 | 3.004E-03 |
|---|
| B cell lineage commitment | GO:0002326 |  | 5.882E-06 | 3.382E-03 |
|---|
| T cell lineage commitment | GO:0002360 |  | 5.882E-06 | 2.706E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.052E-06 | 3.47E-03 |
|---|
| negative regulation of cell development | GO:0010721 |  | 9.052E-06 | 2.974E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 9.052E-06 | 2.602E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.373E-06 | 2.395E-03 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 9.373E-06 | 2.156E-03 |
|---|
| cell fate commitment | GO:0045165 |  | 1.285E-05 | 2.687E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.694E-06 | 3.897E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 3.375E-06 | 3.881E-03 |
|---|
| negative regulation of growth | GO:0045926 |  | 3.918E-06 | 3.004E-03 |
|---|
| B cell lineage commitment | GO:0002326 |  | 5.882E-06 | 3.382E-03 |
|---|
| T cell lineage commitment | GO:0002360 |  | 5.882E-06 | 2.706E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 9.052E-06 | 3.47E-03 |
|---|
| negative regulation of cell development | GO:0010721 |  | 9.052E-06 | 2.974E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 9.052E-06 | 2.602E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.373E-06 | 2.395E-03 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 9.373E-06 | 2.156E-03 |
|---|
| cell fate commitment | GO:0045165 |  | 1.285E-05 | 2.687E-03 |
|---|


