Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1203
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3318 | 1.614e-03 | 2.900e-05 | 1.672e-01 | 7.824e-09 |
|---|
| IPC | 0.3438 | 3.439e-01 | 1.741e-01 | 9.346e-01 | 5.597e-02 |
|---|
| Loi | 0.4823 | 4.200e-05 | 0.000e+00 | 5.047e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6017 | 1.000e-06 | 0.000e+00 | 1.094e-01 | 0.000e+00 |
|---|
| Wang | 0.2573 | 7.658e-03 | 6.639e-02 | 2.301e-01 | 1.170e-04 |
|---|
Expression data for subnetwork 1203 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
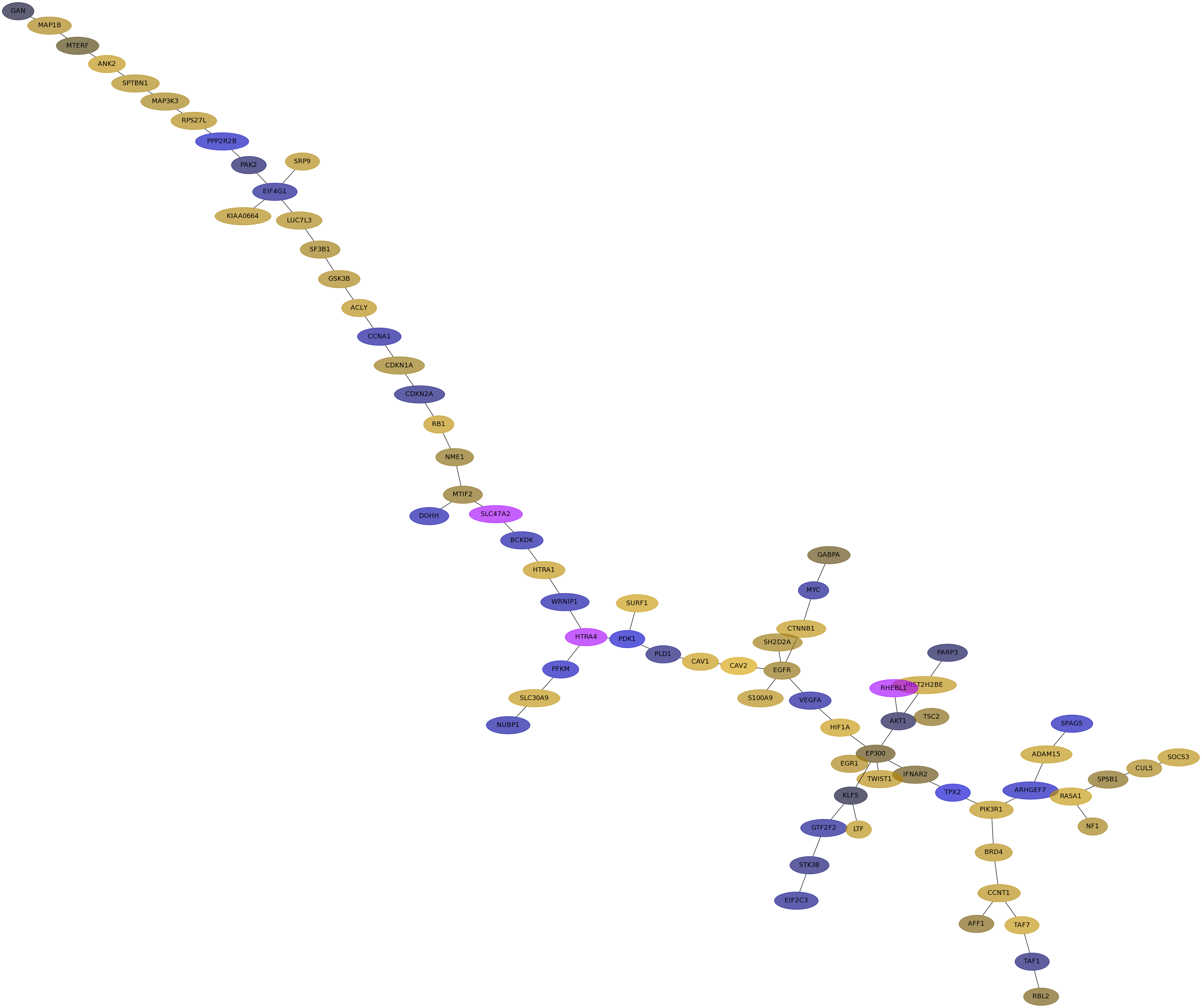
Score for each gene in subnetwork 1203 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| IFNAR2 |   | 4 | 198 | 33 | 41 | 0.021 | -0.100 | 0.055 | 0.106 | -0.058 |
|---|
| TSC2 |   | 3 | 265 | 57 | 71 | 0.046 | 0.236 | 0.044 | 0.114 | -0.102 |
|---|
| HTRA1 |   | 2 | 391 | 492 | 506 | 0.202 | -0.046 | 0.184 | 0.051 | 0.224 |
|---|
| NF1 |   | 1 | 652 | 492 | 517 | 0.097 | 0.110 | -0.052 | -0.032 | -0.094 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| TPX2 |   | 2 | 391 | 492 | 506 | -0.316 | 0.114 | -0.121 | -0.018 | -0.056 |
|---|
| HIST2H2BE |   | 5 | 148 | 492 | 498 | 0.196 | -0.086 | 0.123 | -0.180 | -0.104 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| SPSB1 |   | 7 | 100 | 492 | 495 | 0.044 | -0.124 | 0.088 | -0.152 | 0.169 |
|---|
| LTF |   | 7 | 100 | 492 | 495 | 0.168 | -0.082 | 0.060 | -0.268 | -0.280 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| SPTBN1 |   | 1 | 652 | 492 | 517 | 0.131 | -0.057 | -0.123 | 0.290 | 0.019 |
|---|
| TWIST1 |   | 3 | 265 | 444 | 447 | 0.166 | 0.044 | 0.194 | 0.061 | 0.219 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| DOHH |   | 1 | 652 | 492 | 517 | -0.118 | 0.072 | -0.051 | 0.029 | 0.075 |
|---|
| MAP3K3 |   | 4 | 198 | 206 | 205 | 0.102 | 0.085 | 0.046 | 0.028 | 0.094 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| PDK1 |   | 1 | 652 | 492 | 517 | -0.273 | 0.183 | -0.138 | -0.041 | -0.126 |
|---|
| RPS27L |   | 1 | 652 | 492 | 517 | 0.141 | 0.131 | 0.039 | -0.033 | 0.026 |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| GABPA |   | 6 | 120 | 206 | 200 | 0.019 | 0.033 | -0.183 | 0.066 | -0.161 |
|---|
| MTIF2 |   | 4 | 198 | 492 | 499 | 0.047 | 0.013 | 0.029 | -0.088 | 0.031 |
|---|
| ANK2 |   | 1 | 652 | 492 | 517 | 0.200 | -0.073 | -0.023 | 0.095 | 0.095 |
|---|
| CAV1 |   | 10 | 67 | 439 | 428 | 0.251 | -0.092 | 0.124 | 0.079 | 0.119 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| SOCS3 |   | 3 | 265 | 492 | 503 | 0.180 | 0.201 | -0.110 | 0.064 | -0.138 |
|---|
| EIF4G1 |   | 1 | 652 | 492 | 517 | -0.056 | -0.071 | 0.115 | 0.039 | -0.061 |
|---|
| PARP3 |   | 1 | 652 | 492 | 517 | -0.014 | -0.024 | -0.004 | -0.012 | -0.036 |
|---|
| NME1 |   | 7 | 100 | 492 | 495 | 0.054 | 0.267 | 0.106 | 0.046 | -0.035 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| STK38 |   | 1 | 652 | 492 | 517 | -0.030 | 0.057 | 0.040 | 0.058 | 0.178 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| HTRA4 |   | 3 | 265 | 492 | 503 | undef | 0.102 | undef | undef | undef |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| SLC47A2 |   | 3 | 265 | 492 | 503 | undef | undef | 0.083 | undef | undef |
|---|
| RHEBL1 |   | 2 | 391 | 492 | 506 | undef | 0.154 | undef | undef | undef |
|---|
| SLC30A9 |   | 1 | 652 | 492 | 517 | 0.211 | 0.089 | -0.011 | 0.026 | 0.050 |
|---|
| BCKDK |   | 2 | 391 | 492 | 506 | -0.100 | 0.166 | -0.004 | -0.012 | -0.001 |
|---|
| MTERF |   | 1 | 652 | 492 | 517 | 0.013 | 0.115 | 0.087 | -0.077 | -0.148 |
|---|
| EGR1 |   | 11 | 59 | 283 | 274 | 0.118 | -0.184 | 0.259 | 0.130 | 0.122 |
|---|
| TAF1 |   | 2 | 391 | 492 | 506 | -0.029 | -0.069 | 0.040 | 0.262 | -0.103 |
|---|
| ARHGEF7 |   | 12 | 55 | 492 | 475 | -0.180 | -0.074 | -0.061 | 0.208 | 0.015 |
|---|
| RBL2 |   | 2 | 391 | 492 | 506 | 0.031 | -0.101 | 0.012 | -0.100 | 0.167 |
|---|
| SPAG5 |   | 4 | 198 | 492 | 499 | -0.159 | 0.132 | -0.243 | 0.031 | 0.084 |
|---|
| WRNIP1 |   | 1 | 652 | 492 | 517 | -0.104 | 0.064 | -0.025 | 0.162 | -0.115 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CUL5 |   | 4 | 198 | 492 | 499 | 0.111 | 0.051 | 0.187 | 0.221 | 0.216 |
|---|
| KIAA0664 |   | 1 | 652 | 492 | 517 | 0.144 | 0.039 | -0.045 | -0.014 | 0.031 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| GAN |   | 2 | 391 | 444 | 458 | -0.005 | 0.145 | 0.250 | -0.086 | -0.026 |
|---|
| GTF2F2 |   | 3 | 265 | 283 | 292 | -0.060 | -0.059 | -0.085 | 0.119 | -0.054 |
|---|
| ADAM15 |   | 13 | 47 | 492 | 474 | 0.205 | -0.048 | -0.078 | -0.208 | -0.048 |
|---|
| AFF1 |   | 1 | 652 | 492 | 517 | 0.039 | 0.110 | 0.059 | -0.063 | 0.147 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| SH2D2A |   | 1 | 652 | 492 | 517 | 0.087 | 0.080 | -0.215 | -0.224 | -0.009 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| NUBP1 |   | 1 | 652 | 492 | 517 | -0.096 | 0.140 | -0.191 | 0.057 | 0.073 |
|---|
| PFKM |   | 4 | 198 | 492 | 499 | -0.193 | -0.030 | -0.079 | 0.293 | 0.118 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| RASA1 |   | 9 | 77 | 492 | 494 | 0.237 | -0.079 | 0.146 | 0.127 | 0.149 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| SRP9 |   | 1 | 652 | 492 | 517 | 0.149 | 0.151 | 0.120 | -0.001 | -0.016 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| SURF1 |   | 1 | 652 | 492 | 517 | 0.266 | 0.074 | 0.042 | -0.052 | 0.070 |
|---|
| ACLY |   | 2 | 391 | 412 | 423 | 0.167 | -0.236 | 0.071 | 0.009 | 0.115 |
|---|
| TAF7 |   | 2 | 391 | 492 | 506 | 0.232 | 0.087 | 0.101 | -0.036 | -0.030 |
|---|
| PLD1 |   | 2 | 391 | 492 | 506 | -0.031 | 0.132 | 0.033 | -0.158 | 0.123 |
|---|
| CDKN2A |   | 10 | 67 | 364 | 351 | -0.037 | 0.263 | -0.113 | 0.111 | -0.105 |
|---|
| EIF2C3 |   | 2 | 391 | 206 | 220 | -0.053 | -0.198 | 0.070 | 0.040 | -0.125 |
|---|
GO Enrichment output for subnetwork 1203 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.289E-10 | 5.264E-07 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.693E-10 | 3.097E-07 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 4.308E-10 | 3.303E-07 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.367E-08 | 7.859E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.985E-08 | 9.133E-06 |
|---|
| gas homeostasis | GO:0033483 |  | 3.577E-08 | 1.371E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.651E-07 | 5.425E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.705E-07 | 4.902E-05 |
|---|
| interphase | GO:0051325 |  | 2.041E-07 | 5.216E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.357E-07 | 5.421E-05 |
|---|
| vasculogenesis | GO:0001570 |  | 3.958E-07 | 8.276E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.456E-10 | 3.556E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.631E-10 | 1.993E-07 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.663E-10 | 1.354E-07 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.773E-10 | 1.694E-07 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.229E-09 | 1.089E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.583E-09 | 2.273E-06 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 6.382E-09 | 2.227E-06 |
|---|
| gas homeostasis | GO:0033483 |  | 5.799E-08 | 1.771E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.497E-08 | 2.306E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.039E-07 | 2.539E-05 |
|---|
| G1 phase | GO:0051318 |  | 1.095E-07 | 2.431E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.868E-10 | 4.494E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.113E-10 | 2.542E-07 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.142E-10 | 1.718E-07 |
|---|
| interphase | GO:0051325 |  | 2.886E-10 | 1.736E-07 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.627E-10 | 1.746E-07 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.092E-09 | 8.389E-07 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 7.134E-09 | 2.452E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.426E-09 | 2.534E-06 |
|---|
| G1 phase | GO:0051318 |  | 6.617E-08 | 1.769E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.782E-08 | 2.113E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.326E-07 | 2.9E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.289E-10 | 5.264E-07 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.693E-10 | 3.097E-07 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 4.308E-10 | 3.303E-07 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.367E-08 | 7.859E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.985E-08 | 9.133E-06 |
|---|
| gas homeostasis | GO:0033483 |  | 3.577E-08 | 1.371E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.651E-07 | 5.425E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.705E-07 | 4.902E-05 |
|---|
| interphase | GO:0051325 |  | 2.041E-07 | 5.216E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.357E-07 | 5.421E-05 |
|---|
| vasculogenesis | GO:0001570 |  | 3.958E-07 | 8.276E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.289E-10 | 5.264E-07 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.693E-10 | 3.097E-07 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 4.308E-10 | 3.303E-07 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.367E-08 | 7.859E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.985E-08 | 9.133E-06 |
|---|
| gas homeostasis | GO:0033483 |  | 3.577E-08 | 1.371E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.651E-07 | 5.425E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.705E-07 | 4.902E-05 |
|---|
| interphase | GO:0051325 |  | 2.041E-07 | 5.216E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.357E-07 | 5.421E-05 |
|---|
| vasculogenesis | GO:0001570 |  | 3.958E-07 | 8.276E-05 |
|---|


