Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1202
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3314 | 1.643e-03 | 3.000e-05 | 1.685e-01 | 8.304e-09 |
|---|
| IPC | 0.3438 | 3.438e-01 | 1.741e-01 | 9.346e-01 | 5.596e-02 |
|---|
| Loi | 0.4823 | 4.200e-05 | 0.000e+00 | 5.050e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6022 | 1.000e-06 | 0.000e+00 | 1.083e-01 | 0.000e+00 |
|---|
| Wang | 0.2575 | 7.591e-03 | 6.603e-02 | 2.292e-01 | 1.149e-04 |
|---|
Expression data for subnetwork 1202 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
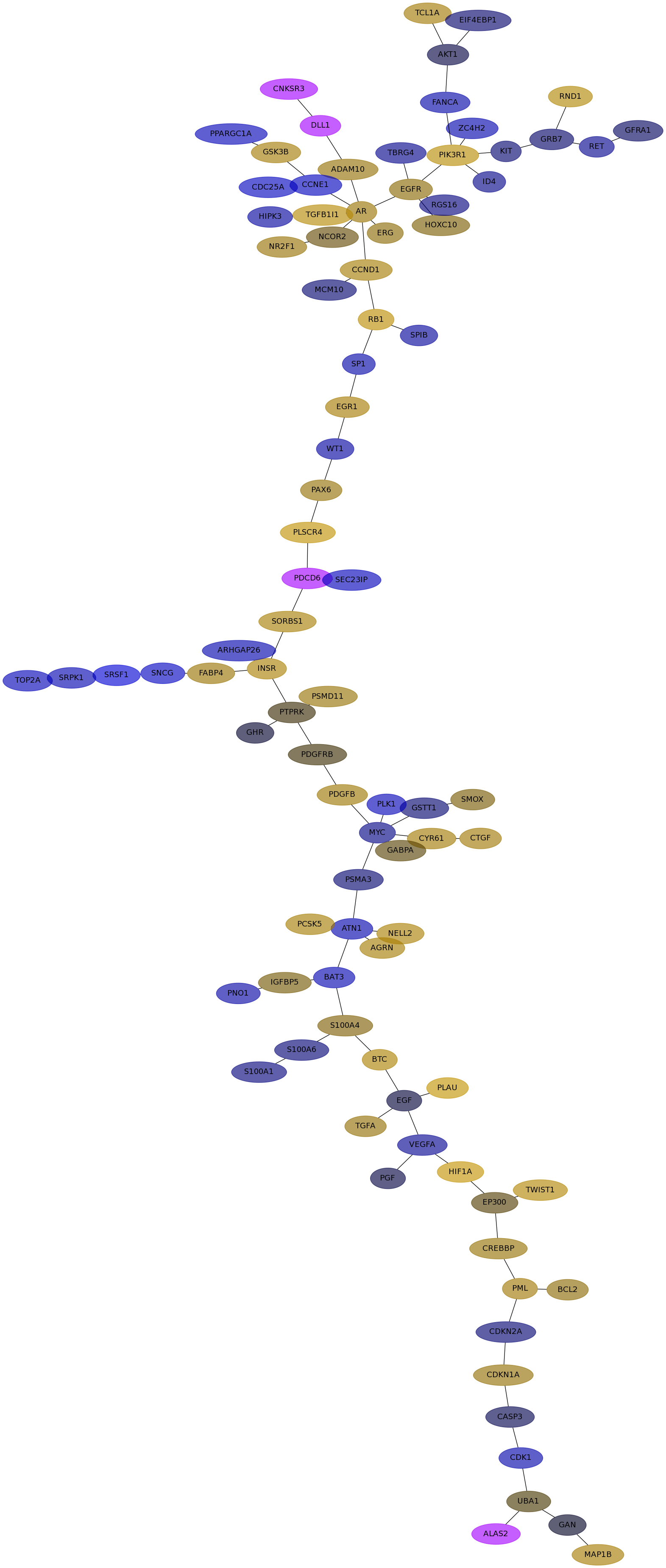
Score for each gene in subnetwork 1202 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| ARHGAP26 |   | 5 | 148 | 444 | 440 | -0.145 | -0.023 | 0.171 | -0.222 | -0.348 |
|---|
| BTC |   | 3 | 265 | 398 | 404 | 0.139 | 0.226 | 0.025 | -0.097 | -0.059 |
|---|
| CASP3 |   | 4 | 198 | 129 | 130 | -0.015 | 0.234 | -0.032 | -0.142 | -0.084 |
|---|
| PAX6 |   | 11 | 59 | 283 | 274 | 0.081 | 0.259 | -0.003 | 0.112 | 0.168 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| PSMA3 |   | 2 | 391 | 444 | 458 | -0.033 | 0.116 | -0.149 | -0.008 | 0.014 |
|---|
| UBA1 |   | 1 | 652 | 444 | 476 | 0.013 | -0.109 | 0.204 | -0.166 | -0.169 |
|---|
| AGRN |   | 1 | 652 | 444 | 476 | 0.121 | -0.055 | 0.341 | -0.106 | -0.024 |
|---|
| ADAM10 |   | 1 | 652 | 444 | 476 | 0.080 | -0.036 | 0.069 | 0.151 | -0.086 |
|---|
| RET |   | 4 | 198 | 57 | 68 | -0.069 | 0.098 | 0.063 | -0.179 | -0.159 |
|---|
| EGF |   | 11 | 59 | 398 | 383 | -0.009 | -0.212 | 0.182 | -0.276 | 0.005 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| PDGFB |   | 2 | 391 | 444 | 458 | 0.100 | 0.020 | 0.046 | 0.085 | 0.338 |
|---|
| CCNE1 |   | 1 | 652 | 444 | 476 | -0.208 | 0.245 | -0.141 | -0.077 | -0.018 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| CREBBP |   | 5 | 148 | 444 | 440 | 0.084 | -0.104 | 0.102 | 0.093 | 0.002 |
|---|
| NELL2 |   | 2 | 391 | 444 | 458 | 0.134 | -0.005 | 0.138 | 0.421 | 0.074 |
|---|
| DLL1 |   | 1 | 652 | 444 | 476 | undef | -0.156 | 0.068 | undef | undef |
|---|
| PLAU |   | 6 | 120 | 444 | 438 | 0.238 | 0.187 | 0.153 | 0.141 | 0.192 |
|---|
| CTGF |   | 10 | 67 | 57 | 52 | 0.106 | -0.169 | 0.181 | 0.126 | 0.209 |
|---|
| CNKSR3 |   | 1 | 652 | 444 | 476 | undef | 0.083 | -0.132 | undef | undef |
|---|
| TWIST1 |   | 3 | 265 | 444 | 447 | 0.166 | 0.044 | 0.194 | 0.061 | 0.219 |
|---|
| BAT3 |   | 2 | 391 | 444 | 458 | -0.153 | -0.078 | 0.072 | -0.057 | 0.069 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| PDGFRB |   | 3 | 265 | 262 | 270 | 0.009 | -0.094 | 0.224 | 0.127 | 0.189 |
|---|
| HOXC10 |   | 4 | 198 | 444 | 444 | 0.044 | -0.015 | 0.116 | -0.133 | -0.017 |
|---|
| PTPRK |   | 6 | 120 | 148 | 156 | 0.008 | 0.156 | 0.141 | -0.094 | -0.024 |
|---|
| PLK1 |   | 5 | 148 | 444 | 440 | -0.179 | 0.142 | -0.143 | 0.094 | -0.163 |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| GABPA |   | 6 | 120 | 206 | 200 | 0.019 | 0.033 | -0.183 | 0.066 | -0.161 |
|---|
| S100A4 |   | 6 | 120 | 283 | 282 | 0.047 | 0.163 | -0.116 | 0.096 | 0.148 |
|---|
| TGFA |   | 8 | 84 | 439 | 429 | 0.077 | -0.057 | -0.027 | -0.158 | 0.020 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| SRSF1 |   | 1 | 652 | 444 | 476 | -0.353 | 0.065 | -0.077 | 0.127 | -0.085 |
|---|
| PGF |   | 1 | 652 | 444 | 476 | -0.011 | -0.012 | 0.059 | 0.005 | 0.072 |
|---|
| EIF4EBP1 |   | 3 | 265 | 444 | 447 | -0.030 | 0.086 | -0.201 | -0.147 | 0.216 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| TCL1A |   | 1 | 652 | 444 | 476 | 0.111 | -0.152 | -0.166 | 0.090 | -0.066 |
|---|
| PPARGC1A |   | 2 | 391 | 444 | 458 | -0.177 | 0.066 | -0.123 | 0.243 | 0.112 |
|---|
| SRPK1 |   | 1 | 652 | 444 | 476 | -0.162 | 0.085 | -0.010 | 0.143 | 0.191 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| FABP4 |   | 3 | 265 | 444 | 447 | 0.091 | -0.036 | -0.028 | 0.227 | 0.107 |
|---|
| PCSK5 |   | 1 | 652 | 444 | 476 | 0.122 | 0.021 | 0.074 | 0.121 | 0.092 |
|---|
| SNCG |   | 1 | 652 | 444 | 476 | -0.226 | 0.080 | -0.086 | 0.088 | -0.226 |
|---|
| RGS16 |   | 2 | 391 | 444 | 458 | -0.048 | 0.202 | 0.009 | -0.004 | 0.283 |
|---|
| NCOR2 |   | 11 | 59 | 283 | 274 | 0.024 | -0.096 | 0.211 | -0.163 | 0.114 |
|---|
| PLSCR4 |   | 2 | 391 | 444 | 458 | 0.223 | -0.069 | 0.140 | 0.231 | 0.212 |
|---|
| MCM10 |   | 3 | 265 | 444 | 447 | -0.033 | 0.178 | -0.245 | 0.015 | -0.063 |
|---|
| EGR1 |   | 11 | 59 | 283 | 274 | 0.118 | -0.184 | 0.259 | 0.130 | 0.122 |
|---|
| GSTT1 |   | 6 | 120 | 364 | 355 | -0.034 | 0.148 | 0.107 | -0.190 | -0.064 |
|---|
| SPIB |   | 1 | 652 | 444 | 476 | -0.094 | -0.034 | -0.202 | -0.070 | -0.179 |
|---|
| PNO1 |   | 1 | 652 | 444 | 476 | -0.118 | 0.090 | -0.052 | -0.092 | -0.138 |
|---|
| RND1 |   | 2 | 391 | 444 | 458 | 0.158 | -0.046 | 0.085 | 0.022 | 0.113 |
|---|
| TOP2A |   | 2 | 391 | 444 | 458 | -0.185 | 0.193 | -0.168 | 0.054 | 0.071 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CYR61 |   | 2 | 391 | 57 | 80 | 0.109 | 0.018 | 0.206 | 0.203 | 0.234 |
|---|
| SMOX |   | 2 | 391 | 444 | 458 | 0.040 | -0.232 | 0.276 | -0.089 | 0.023 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| GAN |   | 2 | 391 | 444 | 458 | -0.005 | 0.145 | 0.250 | -0.086 | -0.026 |
|---|
| WT1 |   | 8 | 84 | 283 | 278 | -0.106 | 0.254 | -0.039 | 0.036 | 0.172 |
|---|
| PSMD11 |   | 2 | 391 | 444 | 458 | 0.085 | 0.088 | -0.090 | -0.137 | -0.078 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| GFRA1 |   | 2 | 391 | 57 | 80 | -0.023 | 0.011 | -0.067 | 0.292 | 0.029 |
|---|
| S100A6 |   | 1 | 652 | 444 | 476 | -0.039 | 0.268 | 0.028 | 0.110 | 0.058 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| CDC25A |   | 5 | 148 | 444 | 440 | -0.220 | 0.113 | -0.170 | 0.151 | -0.003 |
|---|
| ERG |   | 2 | 391 | 444 | 458 | 0.065 | -0.134 | -0.026 | 0.235 | 0.182 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| TBRG4 |   | 2 | 391 | 444 | 458 | -0.066 | 0.173 | -0.060 | -0.073 | -0.055 |
|---|
| ALAS2 |   | 1 | 652 | 444 | 476 | undef | -0.113 | 0.057 | undef | undef |
|---|
| FANCA |   | 3 | 265 | 57 | 71 | -0.131 | 0.120 | -0.188 | 0.106 | -0.053 |
|---|
| NR2F1 |   | 6 | 120 | 444 | 438 | 0.086 | -0.252 | 0.113 | 0.133 | 0.207 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| PDCD6 |   | 4 | 198 | 444 | 444 | undef | 0.091 | -0.060 | undef | undef |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| SEC23IP |   | 1 | 652 | 444 | 476 | -0.194 | 0.033 | -0.006 | -0.033 | -0.055 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| IGFBP5 |   | 3 | 265 | 444 | 447 | 0.038 | -0.076 | 0.110 | 0.074 | 0.181 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| S100A1 |   | 3 | 265 | 283 | 292 | -0.045 | 0.244 | 0.044 | 0.013 | 0.063 |
|---|
| PML |   | 1 | 652 | 444 | 476 | 0.106 | -0.068 | -0.010 | -0.007 | -0.070 |
|---|
| AR |   | 9 | 77 | 283 | 277 | 0.089 | -0.163 | 0.081 | -0.035 | -0.030 |
|---|
| ATN1 |   | 2 | 391 | 444 | 458 | -0.149 | -0.005 | -0.093 | 0.005 | -0.136 |
|---|
| HIPK3 |   | 2 | 391 | 283 | 301 | -0.103 | 0.069 | 0.005 | -0.027 | 0.175 |
|---|
| CDKN2A |   | 10 | 67 | 364 | 351 | -0.037 | 0.263 | -0.113 | 0.111 | -0.105 |
|---|
GO Enrichment output for subnetwork 1202 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.89E-12 | 1.125E-08 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.868E-10 | 4.448E-07 |
|---|
| interphase | GO:0051325 |  | 4.981E-10 | 3.819E-07 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 8.319E-10 | 4.783E-07 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.992E-09 | 9.162E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 9.414E-09 | 3.609E-06 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.39E-08 | 4.566E-06 |
|---|
| response to hypoxia | GO:0001666 |  | 1.974E-08 | 5.674E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.977E-08 | 5.053E-06 |
|---|
| response to oxygen levels | GO:0070482 |  | 2.523E-08 | 5.803E-06 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.667E-07 | 3.486E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.672E-13 | 8.97E-10 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.693E-11 | 5.732E-08 |
|---|
| response to hypoxia | GO:0001666 |  | 1.388E-10 | 1.13E-07 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.429E-10 | 8.726E-08 |
|---|
| response to oxygen levels | GO:0070482 |  | 1.777E-10 | 8.681E-08 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 4.123E-10 | 1.679E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.536E-09 | 8.851E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.541E-09 | 1.081E-06 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.624E-08 | 7.124E-06 |
|---|
| response to UV | GO:0009411 |  | 6.959E-08 | 1.7E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.192E-07 | 2.648E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.076E-13 | 2.184E-09 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.773E-11 | 1.055E-07 |
|---|
| interphase | GO:0051325 |  | 1.236E-10 | 9.909E-08 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.722E-10 | 1.637E-07 |
|---|
| response to hypoxia | GO:0001666 |  | 3.366E-10 | 1.62E-07 |
|---|
| response to oxygen levels | GO:0070482 |  | 4.305E-10 | 1.726E-07 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 7.98E-10 | 2.743E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.401E-09 | 1.324E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.245E-09 | 1.937E-06 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.921E-08 | 1.184E-05 |
|---|
| response to UV | GO:0009411 |  | 9.996E-08 | 2.186E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.89E-12 | 1.125E-08 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.868E-10 | 4.448E-07 |
|---|
| interphase | GO:0051325 |  | 4.981E-10 | 3.819E-07 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 8.319E-10 | 4.783E-07 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.992E-09 | 9.162E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 9.414E-09 | 3.609E-06 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.39E-08 | 4.566E-06 |
|---|
| response to hypoxia | GO:0001666 |  | 1.974E-08 | 5.674E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.977E-08 | 5.053E-06 |
|---|
| response to oxygen levels | GO:0070482 |  | 2.523E-08 | 5.803E-06 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.667E-07 | 3.486E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.89E-12 | 1.125E-08 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.868E-10 | 4.448E-07 |
|---|
| interphase | GO:0051325 |  | 4.981E-10 | 3.819E-07 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 8.319E-10 | 4.783E-07 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.992E-09 | 9.162E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 9.414E-09 | 3.609E-06 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.39E-08 | 4.566E-06 |
|---|
| response to hypoxia | GO:0001666 |  | 1.974E-08 | 5.674E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.977E-08 | 5.053E-06 |
|---|
| response to oxygen levels | GO:0070482 |  | 2.523E-08 | 5.803E-06 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.667E-07 | 3.486E-05 |
|---|


