Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1155
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3327 | 1.550e-03 | 2.800e-05 | 1.643e-01 | 7.130e-09 |
|---|
| IPC | 0.3411 | 3.484e-01 | 1.780e-01 | 9.356e-01 | 5.803e-02 |
|---|
| Loi | 0.4768 | 5.400e-05 | 0.000e+00 | 5.320e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5995 | 1.000e-06 | 0.000e+00 | 1.139e-01 | 0.000e+00 |
|---|
| Wang | 0.2570 | 7.733e-03 | 6.680e-02 | 2.311e-01 | 1.194e-04 |
|---|
Expression data for subnetwork 1155 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
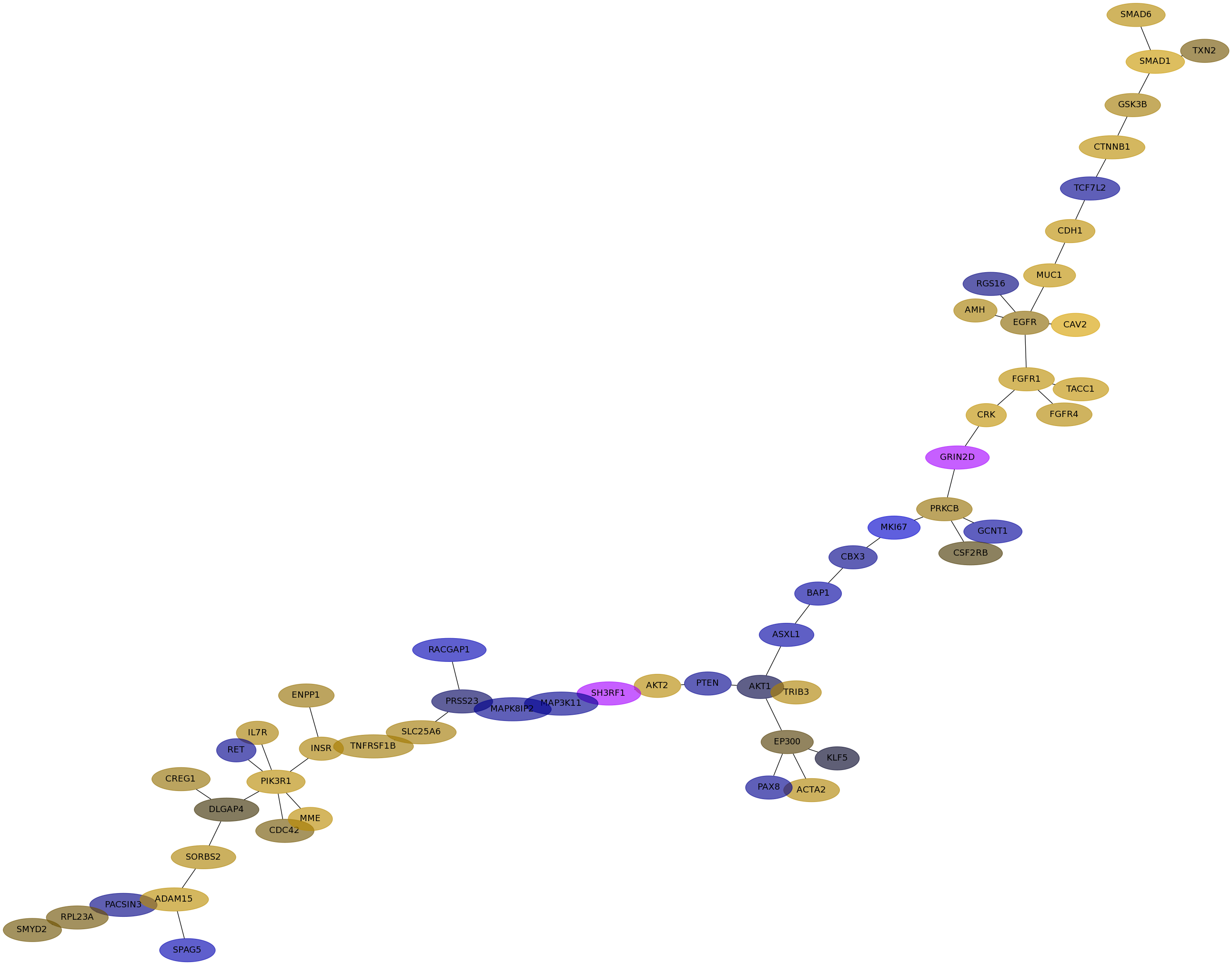
Score for each gene in subnetwork 1155 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| RGS16 |   | 2 | 391 | 444 | 458 | -0.048 | 0.202 | 0.009 | -0.004 | 0.283 |
|---|
| RET |   | 4 | 198 | 57 | 68 | -0.069 | 0.098 | 0.063 | -0.179 | -0.159 |
|---|
| DLGAP4 |   | 5 | 148 | 326 | 330 | 0.010 | -0.029 | 0.180 | -0.201 | -0.043 |
|---|
| RACGAP1 |   | 3 | 265 | 778 | 770 | -0.166 | 0.283 | -0.164 | 0.073 | 0.188 |
|---|
| SPAG5 |   | 4 | 198 | 492 | 499 | -0.159 | 0.132 | -0.243 | 0.031 | 0.084 |
|---|
| RPL23A |   | 10 | 67 | 57 | 52 | 0.031 | 0.208 | -0.007 | 0.024 | 0.055 |
|---|
| CSF2RB |   | 3 | 265 | 691 | 685 | 0.013 | 0.013 | -0.196 | -0.109 | -0.141 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| PTEN |   | 1 | 652 | 778 | 795 | -0.070 | 0.045 | 0.055 | 0.017 | -0.041 |
|---|
| AMH |   | 3 | 265 | 778 | 770 | 0.124 | 0.103 | 0.056 | 0.113 | 0.301 |
|---|
| ACTA2 |   | 2 | 391 | 778 | 789 | 0.155 | -0.147 | 0.256 | 0.125 | 0.220 |
|---|
| ADAM15 |   | 13 | 47 | 492 | 474 | 0.205 | -0.048 | -0.078 | -0.208 | -0.048 |
|---|
| SMAD6 |   | 4 | 198 | 665 | 650 | 0.173 | -0.241 | 0.053 | 0.266 | 0.070 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| GCNT1 |   | 2 | 391 | 778 | 789 | -0.091 | 0.080 | 0.137 | 0.013 | 0.014 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| PRSS23 |   | 2 | 391 | 778 | 789 | -0.023 | 0.133 | 0.056 | 0.060 | 0.092 |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| TACC1 |   | 5 | 148 | 326 | 330 | 0.229 | -0.051 | 0.020 | -0.022 | 0.126 |
|---|
| CREG1 |   | 5 | 148 | 691 | 670 | 0.080 | 0.029 | 0.018 | -0.189 | -0.176 |
|---|
| ASXL1 |   | 1 | 652 | 778 | 795 | -0.114 | 0.044 | 0.146 | 0.031 | 0.041 |
|---|
| AKT2 |   | 3 | 265 | 778 | 770 | 0.179 | 0.110 | -0.118 | -0.003 | -0.092 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PACSIN3 |   | 3 | 265 | 778 | 770 | -0.055 | 0.270 | 0.129 | -0.045 | 0.066 |
|---|
| MKI67 |   | 3 | 265 | 778 | 770 | -0.295 | 0.205 | -0.123 | 0.176 | 0.018 |
|---|
| SORBS2 |   | 3 | 265 | 778 | 770 | 0.147 | -0.075 | 0.077 | 0.084 | -0.114 |
|---|
| BAP1 |   | 1 | 652 | 778 | 795 | -0.106 | -0.077 | -0.003 | 0.070 | 0.046 |
|---|
| SMYD2 |   | 4 | 198 | 148 | 162 | 0.032 | 0.260 | 0.068 | 0.179 | 0.191 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| MAPK8IP2 |   | 1 | 652 | 778 | 795 | -0.071 | 0.141 | -0.097 | -0.029 | -0.144 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| CBX3 |   | 1 | 652 | 778 | 795 | -0.065 | 0.201 | 0.019 | 0.026 | 0.011 |
|---|
| TNFRSF1B |   | 12 | 55 | 563 | 544 | 0.109 | 0.024 | -0.187 | -0.096 | -0.106 |
|---|
| TRIB3 |   | 1 | 652 | 778 | 795 | 0.149 | 0.119 | 0.133 | -0.046 | 0.093 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| ENPP1 |   | 3 | 265 | 778 | 770 | 0.082 | 0.088 | 0.129 | -0.026 | 0.039 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| PAX8 |   | 4 | 198 | 778 | 766 | -0.072 | 0.008 | -0.083 | 0.032 | -0.002 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| PRKCB |   | 8 | 84 | 33 | 35 | 0.081 | -0.065 | 0.001 | -0.148 | -0.035 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| MAP3K11 |   | 1 | 652 | 778 | 795 | -0.071 | 0.001 | -0.090 | 0.073 | -0.048 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| TXN2 |   | 4 | 198 | 33 | 41 | 0.035 | -0.112 | -0.009 | -0.044 | -0.139 |
|---|
| MME |   | 8 | 84 | 57 | 54 | 0.200 | -0.121 | 0.020 | 0.088 | -0.130 |
|---|
| SLC25A6 |   | 3 | 265 | 778 | 770 | 0.111 | -0.178 | 0.115 | 0.040 | -0.080 |
|---|
GO Enrichment output for subnetwork 1155 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.443E-08 | 1.942E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.025E-07 | 1.178E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.72E-06 | 1.319E-03 |
|---|
| regulation of cellular component biogenesis | GO:0044087 |  | 1.772E-06 | 1.019E-03 |
|---|
| nuclear migration | GO:0007097 |  | 2.087E-06 | 9.601E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.087E-06 | 8.001E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.087E-06 | 6.858E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.753E-06 | 7.916E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 4.156E-06 | 1.062E-03 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 4.156E-06 | 9.559E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 5.409E-06 | 1.131E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.617E-12 | 1.128E-08 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 2.399E-09 | 2.93E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 7.17E-09 | 5.839E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.667E-08 | 1.018E-05 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.321E-08 | 1.623E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.253E-08 | 1.732E-05 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 9.89E-08 | 3.452E-05 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.548E-07 | 4.729E-05 |
|---|
| negative regulation of protein kinase cascade | GO:0010741 |  | 2.878E-07 | 7.811E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.03E-07 | 7.402E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 9.831E-07 | 2.183E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.09E-12 | 7.435E-09 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 3.106E-09 | 3.736E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 9.281E-09 | 7.444E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 2.157E-08 | 1.298E-05 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.297E-08 | 2.068E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.47E-08 | 1.793E-05 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.279E-07 | 4.396E-05 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.002E-07 | 6.021E-05 |
|---|
| negative regulation of protein kinase cascade | GO:0010741 |  | 3.945E-07 | 1.055E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.392E-07 | 1.057E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.173E-06 | 2.565E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.443E-08 | 1.942E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.025E-07 | 1.178E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.72E-06 | 1.319E-03 |
|---|
| regulation of cellular component biogenesis | GO:0044087 |  | 1.772E-06 | 1.019E-03 |
|---|
| nuclear migration | GO:0007097 |  | 2.087E-06 | 9.601E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.087E-06 | 8.001E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.087E-06 | 6.858E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.753E-06 | 7.916E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 4.156E-06 | 1.062E-03 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 4.156E-06 | 9.559E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 5.409E-06 | 1.131E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.443E-08 | 1.942E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.025E-07 | 1.178E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.72E-06 | 1.319E-03 |
|---|
| regulation of cellular component biogenesis | GO:0044087 |  | 1.772E-06 | 1.019E-03 |
|---|
| nuclear migration | GO:0007097 |  | 2.087E-06 | 9.601E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.087E-06 | 8.001E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.087E-06 | 6.858E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.753E-06 | 7.916E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 4.156E-06 | 1.062E-03 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 4.156E-06 | 9.559E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 5.409E-06 | 1.131E-03 |
|---|


