Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1154
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3323 | 1.580e-03 | 2.800e-05 | 1.656e-01 | 7.328e-09 |
|---|
| IPC | 0.3410 | 3.485e-01 | 1.781e-01 | 9.356e-01 | 5.808e-02 |
|---|
| Loi | 0.4767 | 5.400e-05 | 0.000e+00 | 5.324e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6001 | 1.000e-06 | 0.000e+00 | 1.125e-01 | 0.000e+00 |
|---|
| Wang | 0.2572 | 7.668e-03 | 6.645e-02 | 2.302e-01 | 1.173e-04 |
|---|
Expression data for subnetwork 1154 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
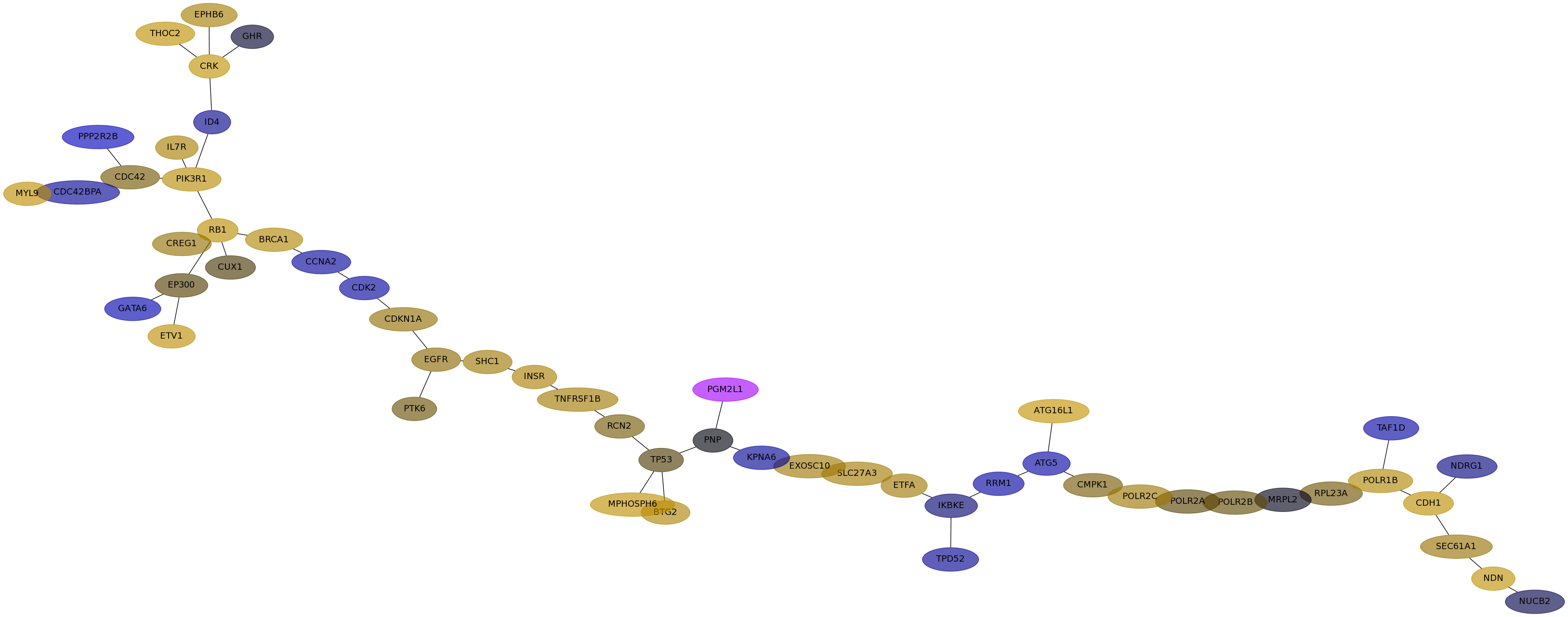
Score for each gene in subnetwork 1154 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| MYL9 |   | 1 | 652 | 756 | 780 | 0.212 | -0.087 | 0.159 | 0.174 | 0.152 |
|---|
| PTK6 |   | 2 | 391 | 206 | 220 | 0.029 | -0.010 | 0.204 | -0.343 | 0.213 |
|---|
| NDRG1 |   | 3 | 265 | 756 | 748 | -0.049 | 0.222 | 0.143 | -0.040 | 0.242 |
|---|
| RRM1 |   | 1 | 652 | 756 | 780 | -0.105 | 0.258 | 0.039 | 0.114 | 0.098 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| NUCB2 |   | 1 | 652 | 756 | 780 | -0.013 | 0.010 | 0.130 | -0.038 | -0.042 |
|---|
| RPL23A |   | 10 | 67 | 57 | 52 | 0.031 | 0.208 | -0.007 | 0.024 | 0.055 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| SHC1 |   | 4 | 198 | 691 | 671 | 0.102 | -0.090 | -0.041 | -0.005 | -0.138 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| ETFA |   | 3 | 265 | 756 | 748 | 0.105 | 0.027 | 0.094 | 0.088 | 0.013 |
|---|
| POLR1B |   | 11 | 59 | 129 | 126 | 0.157 | 0.062 | -0.036 | -0.079 | 0.139 |
|---|
| EPHB6 |   | 7 | 100 | 419 | 407 | 0.122 | 0.008 | 0.042 | -0.219 | -0.059 |
|---|
| CMPK1 |   | 2 | 391 | 756 | 756 | 0.039 | -0.054 | 0.193 | -0.038 | 0.096 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| ETV1 |   | 11 | 59 | 746 | 724 | 0.214 | 0.046 | 0.054 | -0.065 | 0.141 |
|---|
| MPHOSPH6 |   | 8 | 84 | 756 | 726 | 0.217 | -0.084 | 0.175 | -0.269 | 0.146 |
|---|
| POLR2B |   | 2 | 391 | 756 | 756 | 0.024 | -0.064 | 0.012 | 0.095 | -0.072 |
|---|
| KPNA6 |   | 5 | 148 | 182 | 181 | -0.082 | -0.144 | 0.128 | -0.005 | -0.108 |
|---|
| PNP |   | 1 | 652 | 756 | 780 | -0.001 | 0.036 | -0.012 | 0.108 | 0.132 |
|---|
| CREG1 |   | 5 | 148 | 691 | 670 | 0.080 | 0.029 | 0.018 | -0.189 | -0.176 |
|---|
| TAF1D |   | 5 | 148 | 57 | 63 | -0.110 | 0.141 | 0.113 | 0.041 | 0.059 |
|---|
| BTG2 |   | 6 | 120 | 1 | 21 | 0.146 | -0.029 | -0.093 | -0.111 | -0.170 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| ATG16L1 |   | 2 | 391 | 756 | 756 | 0.244 | -0.123 | 0.108 | -0.112 | -0.175 |
|---|
| CUX1 |   | 8 | 84 | 271 | 266 | 0.012 | -0.042 | 0.256 | -0.121 | 0.040 |
|---|
| TPD52 |   | 1 | 652 | 756 | 780 | -0.074 | 0.014 | -0.097 | -0.139 | 0.160 |
|---|
| CDC42BPA |   | 2 | 391 | 756 | 756 | -0.082 | -0.102 | 0.098 | 0.059 | 0.260 |
|---|
| BRCA1 |   | 5 | 148 | 598 | 575 | 0.157 | -0.201 | -0.078 | -0.087 | 0.168 |
|---|
| RCN2 |   | 6 | 120 | 756 | 731 | 0.037 | 0.059 | 0.051 | 0.176 | -0.098 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| EXOSC10 |   | 1 | 652 | 756 | 780 | 0.104 | -0.148 | 0.142 | -0.098 | 0.115 |
|---|
| POLR2A |   | 5 | 148 | 271 | 267 | 0.019 | -0.107 | 0.247 | 0.091 | 0.141 |
|---|
| TNFRSF1B |   | 12 | 55 | 563 | 544 | 0.109 | 0.024 | -0.187 | -0.096 | -0.106 |
|---|
| POLR2C |   | 2 | 391 | 756 | 756 | 0.097 | -0.130 | 0.002 | -0.204 | 0.168 |
|---|
| MRPL2 |   | 1 | 652 | 756 | 780 | -0.003 | -0.035 | -0.053 | -0.090 | 0.095 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| IKBKE |   | 8 | 84 | 756 | 726 | -0.033 | 0.227 | -0.041 | -0.095 | 0.079 |
|---|
| SEC61A1 |   | 5 | 148 | 271 | 267 | 0.085 | -0.161 | 0.128 | -0.096 | 0.070 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| GATA6 |   | 1 | 652 | 756 | 780 | -0.147 | -0.063 | 0.018 | -0.071 | 0.203 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| THOC2 |   | 11 | 59 | 419 | 406 | 0.217 | -0.094 | 0.119 | 0.068 | 0.072 |
|---|
| NDN |   | 3 | 265 | 756 | 748 | 0.229 | -0.097 | 0.169 | 0.083 | 0.245 |
|---|
| PGM2L1 |   | 1 | 652 | 756 | 780 | undef | -0.019 | 0.220 | undef | undef |
|---|
| ATG5 |   | 2 | 391 | 756 | 756 | -0.116 | 0.195 | -0.113 | -0.013 | 0.210 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| SLC27A3 |   | 2 | 391 | 756 | 756 | 0.117 | -0.019 | -0.027 | 0.058 | 0.028 |
|---|
GO Enrichment output for subnetwork 1154 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 4.074E-09 | 9.371E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.828E-08 | 3.253E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.828E-08 | 2.168E-05 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 8.093E-08 | 4.654E-05 |
|---|
| nucleus localization | GO:0051647 |  | 1.01E-07 | 4.645E-05 |
|---|
| cellular response to starvation | GO:0009267 |  | 7.856E-07 | 3.012E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.4E-06 | 4.601E-04 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 1.542E-06 | 4.434E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.542E-06 | 3.942E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 2.362E-06 | 5.432E-04 |
|---|
| response to starvation | GO:0042594 |  | 2.979E-06 | 6.23E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.64E-09 | 8.891E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 5.644E-09 | 6.894E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.688E-08 | 1.375E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.688E-08 | 1.031E-05 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 2.364E-08 | 1.155E-05 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 2.364E-08 | 9.624E-06 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 3.791E-08 | 1.323E-05 |
|---|
| nucleus localization | GO:0051647 |  | 7.883E-08 | 2.407E-05 |
|---|
| leukocyte differentiation | GO:0002521 |  | 1.43E-07 | 3.881E-05 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.482E-07 | 3.62E-05 |
|---|
| lung development | GO:0030324 |  | 3.563E-07 | 7.913E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 5.093E-09 | 1.225E-05 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 6.06E-09 | 7.291E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.184E-08 | 9.499E-06 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 2.121E-08 | 1.276E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.361E-08 | 1.136E-05 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 2.775E-08 | 1.113E-05 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 3.578E-08 | 1.23E-05 |
|---|
| nucleus localization | GO:0051647 |  | 1.102E-07 | 3.314E-05 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.575E-07 | 4.209E-05 |
|---|
| leukocyte differentiation | GO:0002521 |  | 2.166E-07 | 5.211E-05 |
|---|
| lung development | GO:0030324 |  | 4.846E-07 | 1.06E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 4.074E-09 | 9.371E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.828E-08 | 3.253E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.828E-08 | 2.168E-05 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 8.093E-08 | 4.654E-05 |
|---|
| nucleus localization | GO:0051647 |  | 1.01E-07 | 4.645E-05 |
|---|
| cellular response to starvation | GO:0009267 |  | 7.856E-07 | 3.012E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.4E-06 | 4.601E-04 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 1.542E-06 | 4.434E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.542E-06 | 3.942E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 2.362E-06 | 5.432E-04 |
|---|
| response to starvation | GO:0042594 |  | 2.979E-06 | 6.23E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 4.074E-09 | 9.371E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.828E-08 | 3.253E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.828E-08 | 2.168E-05 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 8.093E-08 | 4.654E-05 |
|---|
| nucleus localization | GO:0051647 |  | 1.01E-07 | 4.645E-05 |
|---|
| cellular response to starvation | GO:0009267 |  | 7.856E-07 | 3.012E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.4E-06 | 4.601E-04 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 1.542E-06 | 4.434E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.542E-06 | 3.942E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 2.362E-06 | 5.432E-04 |
|---|
| response to starvation | GO:0042594 |  | 2.979E-06 | 6.23E-04 |
|---|


