Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1152
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3323 | 1.578e-03 | 2.800e-05 | 1.656e-01 | 7.316e-09 |
|---|
| IPC | 0.3412 | 3.482e-01 | 1.779e-01 | 9.355e-01 | 5.795e-02 |
|---|
| Loi | 0.4772 | 5.300e-05 | 0.000e+00 | 5.300e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6003 | 1.000e-06 | 0.000e+00 | 1.121e-01 | 0.000e+00 |
|---|
| Wang | 0.2566 | 7.849e-03 | 6.742e-02 | 2.327e-01 | 1.232e-04 |
|---|
Expression data for subnetwork 1152 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
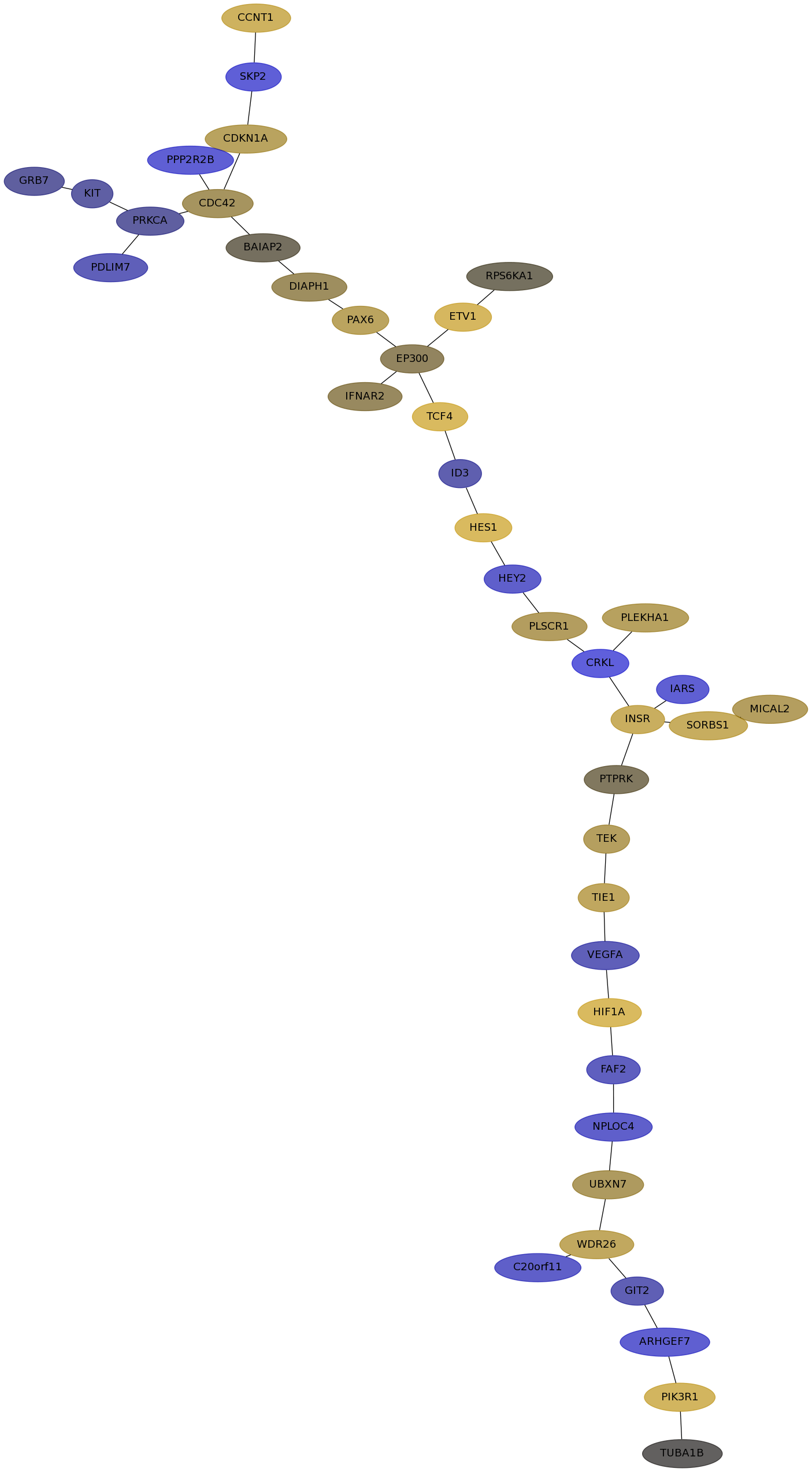
Score for each gene in subnetwork 1152 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| TEK |   | 7 | 100 | 148 | 155 | 0.063 | -0.149 | -0.049 | 0.174 | 0.113 |
|---|
| IFNAR2 |   | 4 | 198 | 33 | 41 | 0.021 | -0.100 | 0.055 | 0.106 | -0.058 |
|---|
| PAX6 |   | 11 | 59 | 283 | 274 | 0.081 | 0.259 | -0.003 | 0.112 | 0.168 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| FAF2 |   | 2 | 391 | 148 | 171 | -0.093 | 0.027 | 0.212 | -0.070 | 0.195 |
|---|
| ARHGEF7 |   | 12 | 55 | 492 | 475 | -0.180 | -0.074 | -0.061 | 0.208 | 0.015 |
|---|
| RPS6KA1 |   | 8 | 84 | 696 | 669 | 0.005 | -0.208 | 0.084 | -0.322 | -0.071 |
|---|
| WDR26 |   | 2 | 391 | 584 | 578 | 0.101 | 0.007 | 0.149 | -0.028 | 0.110 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| PRKCA |   | 5 | 148 | 339 | 337 | -0.029 | -0.016 | -0.088 | 0.174 | 0.005 |
|---|
| GIT2 |   | 3 | 265 | 584 | 573 | -0.065 | -0.191 | 0.362 | -0.164 | 0.054 |
|---|
| NPLOC4 |   | 2 | 391 | 148 | 171 | -0.146 | -0.237 | -0.168 | 0.106 | 0.019 |
|---|
| UBXN7 |   | 6 | 120 | 148 | 156 | 0.049 | -0.055 | 0.153 | 0.224 | 0.175 |
|---|
| HEY2 |   | 2 | 391 | 746 | 751 | -0.147 | -0.045 | -0.021 | 0.398 | -0.058 |
|---|
| CRKL |   | 7 | 100 | 326 | 317 | -0.271 | -0.023 | 0.056 | 0.271 | -0.008 |
|---|
| IARS |   | 1 | 652 | 746 | 767 | -0.195 | -0.141 | 0.033 | -0.030 | -0.056 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| TCF4 |   | 1 | 652 | 746 | 767 | 0.249 | -0.109 | 0.229 | 0.063 | 0.097 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| ETV1 |   | 11 | 59 | 746 | 724 | 0.214 | 0.046 | 0.054 | -0.065 | 0.141 |
|---|
| PTPRK |   | 6 | 120 | 148 | 156 | 0.008 | 0.156 | 0.141 | -0.094 | -0.024 |
|---|
| C20orf11 |   | 1 | 652 | 746 | 767 | -0.135 | 0.078 | -0.131 | 0.073 | 0.034 |
|---|
| PDLIM7 |   | 2 | 391 | 398 | 411 | -0.075 | -0.054 | 0.224 | -0.015 | 0.268 |
|---|
| HES1 |   | 4 | 198 | 746 | 725 | 0.247 | 0.115 | 0.157 | 0.010 | -0.106 |
|---|
| MICAL2 |   | 3 | 265 | 182 | 189 | 0.061 | -0.012 | 0.143 | -0.042 | 0.161 |
|---|
| PLSCR1 |   | 7 | 100 | 634 | 614 | 0.058 | 0.202 | -0.159 | 0.091 | -0.105 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| PLEKHA1 |   | 2 | 391 | 746 | 751 | 0.068 | 0.208 | -0.052 | 0.166 | 0.179 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| DIAPH1 |   | 2 | 391 | 746 | 751 | 0.027 | 0.088 | 0.139 | -0.074 | 0.086 |
|---|
| BAIAP2 |   | 2 | 391 | 746 | 751 | 0.005 | -0.077 | -0.025 | 0.099 | -0.051 |
|---|
| TUBA1B |   | 7 | 100 | 182 | 180 | 0.001 | 0.116 | 0.094 | 0.249 | 0.128 |
|---|
| ID3 |   | 2 | 391 | 746 | 751 | -0.051 | 0.072 | -0.006 | 0.192 | 0.057 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| TIE1 |   | 6 | 120 | 148 | 156 | 0.094 | -0.108 | 0.005 | 0.141 | 0.252 |
|---|
GO Enrichment output for subnetwork 1152 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.421E-08 | 7.869E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 8.981E-08 | 1.033E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 1.595E-07 | 1.223E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.595E-07 | 9.172E-05 |
|---|
| microspike assembly | GO:0030035 |  | 2.384E-07 | 1.097E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 5.662E-07 | 2.171E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.063E-06 | 3.492E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.063E-06 | 3.056E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.063E-06 | 2.716E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.894E-06 | 4.355E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.118E-06 | 4.429E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.398E-08 | 5.858E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.335E-08 | 6.517E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 9.338E-08 | 7.604E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.209E-07 | 1.96E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 4.424E-07 | 2.162E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.21E-07 | 2.121E-04 |
|---|
| microspike assembly | GO:0030035 |  | 5.672E-07 | 1.979E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 9.542E-07 | 2.914E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.039E-06 | 2.821E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.039E-06 | 2.539E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.34E-06 | 2.976E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.875E-08 | 6.917E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.447E-08 | 6.553E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.119E-07 | 8.976E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.486E-07 | 2.097E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 4.065E-07 | 1.956E-04 |
|---|
| microspike assembly | GO:0030035 |  | 5.299E-07 | 2.125E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.98E-07 | 2.055E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.066E-06 | 3.205E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.193E-06 | 3.188E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.193E-06 | 2.869E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.316E-06 | 2.879E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.421E-08 | 7.869E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 8.981E-08 | 1.033E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 1.595E-07 | 1.223E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.595E-07 | 9.172E-05 |
|---|
| microspike assembly | GO:0030035 |  | 2.384E-07 | 1.097E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 5.662E-07 | 2.171E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.063E-06 | 3.492E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.063E-06 | 3.056E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.063E-06 | 2.716E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.894E-06 | 4.355E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.118E-06 | 4.429E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.421E-08 | 7.869E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 8.981E-08 | 1.033E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 1.595E-07 | 1.223E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.595E-07 | 9.172E-05 |
|---|
| microspike assembly | GO:0030035 |  | 2.384E-07 | 1.097E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 5.662E-07 | 2.171E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.063E-06 | 3.492E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.063E-06 | 3.056E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.063E-06 | 2.716E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.894E-06 | 4.355E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.118E-06 | 4.429E-04 |
|---|


