Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1145
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3339 | 1.472e-03 | 2.500e-05 | 1.607e-01 | 5.913e-09 |
|---|
| IPC | 0.3408 | 3.488e-01 | 1.784e-01 | 9.356e-01 | 5.823e-02 |
|---|
| Loi | 0.4762 | 5.500e-05 | 0.000e+00 | 5.351e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5978 | 1.000e-06 | 0.000e+00 | 1.174e-01 | 0.000e+00 |
|---|
| Wang | 0.2572 | 7.684e-03 | 6.653e-02 | 2.305e-01 | 1.178e-04 |
|---|
Expression data for subnetwork 1145 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
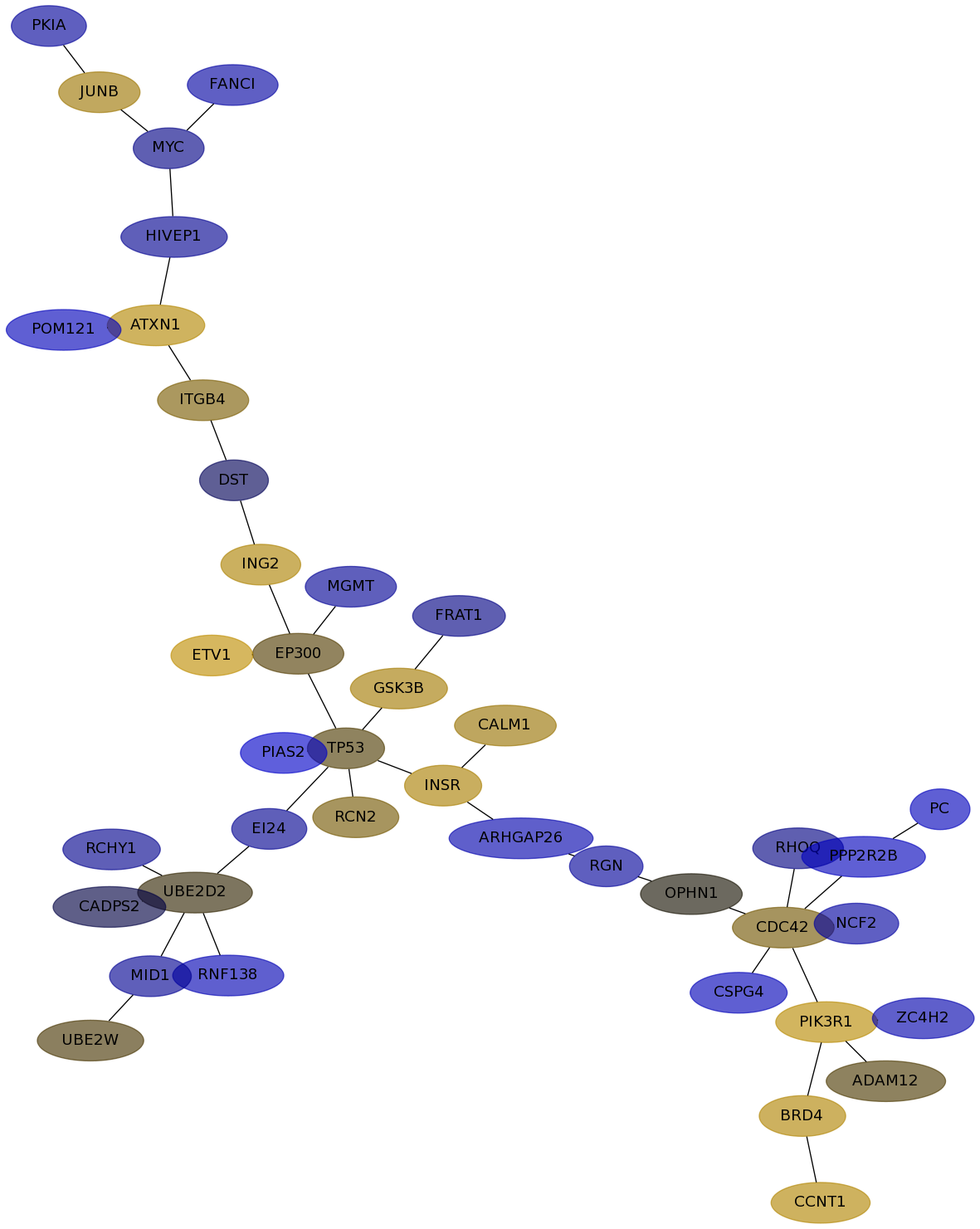
Score for each gene in subnetwork 1145 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| ARHGAP26 |   | 5 | 148 | 444 | 440 | -0.145 | -0.023 | 0.171 | -0.222 | -0.348 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| UBE2W |   | 1 | 652 | 832 | 850 | 0.013 | 0.201 | -0.264 | 0.150 | 0.066 |
|---|
| OPHN1 |   | 5 | 148 | 339 | 337 | 0.003 | 0.062 | 0.278 | 0.194 | -0.182 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| RCHY1 |   | 1 | 652 | 832 | 850 | -0.069 | -0.035 | 0.007 | 0.245 | -0.026 |
|---|
| NCF2 |   | 1 | 652 | 832 | 850 | -0.108 | 0.193 | -0.104 | -0.069 | 0.034 |
|---|
| POM121 |   | 5 | 148 | 419 | 415 | -0.195 | -0.197 | 0.101 | 0.079 | 0.017 |
|---|
| FANCI |   | 4 | 198 | 832 | 809 | -0.111 | 0.145 | -0.194 | 0.104 | -0.036 |
|---|
| RGN |   | 1 | 652 | 832 | 850 | -0.092 | -0.051 | -0.050 | 0.184 | -0.120 |
|---|
| DST |   | 2 | 391 | 419 | 433 | -0.019 | -0.094 | 0.343 | 0.065 | 0.194 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| JUNB |   | 5 | 148 | 696 | 672 | 0.101 | 0.057 | 0.086 | 0.065 | -0.015 |
|---|
| EI24 |   | 8 | 84 | 339 | 336 | -0.074 | 0.051 | 0.078 | 0.107 | 0.070 |
|---|
| CADPS2 |   | 1 | 652 | 832 | 850 | -0.011 | -0.080 | 0.102 | -0.062 | -0.003 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| ETV1 |   | 11 | 59 | 746 | 724 | 0.214 | 0.046 | 0.054 | -0.065 | 0.141 |
|---|
| CSPG4 |   | 2 | 391 | 823 | 817 | -0.188 | 0.051 | 0.088 | 0.190 | -0.027 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| RHOQ |   | 1 | 652 | 832 | 850 | -0.054 | -0.187 | -0.072 | -0.010 | -0.082 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| CALM1 |   | 3 | 265 | 57 | 71 | 0.091 | 0.014 | 0.021 | 0.192 | 0.247 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| ING2 |   | 1 | 652 | 832 | 850 | 0.147 | 0.189 | 0.168 | 0.051 | -0.054 |
|---|
| ATXN1 |   | 6 | 120 | 364 | 355 | 0.171 | 0.026 | 0.192 | 0.175 | 0.163 |
|---|
| ADAM12 |   | 2 | 391 | 832 | 831 | 0.015 | -0.108 | 0.093 | -0.016 | 0.201 |
|---|
| RCN2 |   | 6 | 120 | 756 | 731 | 0.037 | 0.059 | 0.051 | 0.176 | -0.098 |
|---|
| MGMT |   | 2 | 391 | 832 | 831 | -0.082 | 0.184 | -0.022 | 0.201 | 0.196 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| PC |   | 1 | 652 | 832 | 850 | -0.201 | -0.171 | -0.017 | -0.024 | -0.060 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| FRAT1 |   | 2 | 391 | 832 | 831 | -0.055 | -0.013 | -0.125 | 0.156 | 0.078 |
|---|
| PIAS2 |   | 3 | 265 | 620 | 613 | -0.270 | 0.039 | -0.072 | 0.194 | -0.172 |
|---|
| UBE2D2 |   | 2 | 391 | 536 | 545 | 0.007 | 0.212 | 0.002 | 0.217 | 0.090 |
|---|
| RNF138 |   | 1 | 652 | 832 | 850 | -0.168 | 0.021 | -0.180 | 0.167 | 0.049 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| ITGB4 |   | 6 | 120 | 364 | 355 | 0.044 | 0.021 | 0.313 | -0.134 | -0.114 |
|---|
| MID1 |   | 1 | 652 | 832 | 850 | -0.078 | -0.047 | 0.156 | 0.068 | 0.166 |
|---|
| PKIA |   | 3 | 265 | 832 | 816 | -0.091 | -0.120 | 0.085 | 0.249 | 0.044 |
|---|
| HIVEP1 |   | 6 | 120 | 419 | 410 | -0.072 | -0.199 | 0.069 | 0.000 | -0.042 |
|---|
GO Enrichment output for subnetwork 1145 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 8.49E-07 | 1.953E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 8.49E-07 | 9.763E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.693E-06 | 1.298E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.693E-06 | 9.732E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.952E-06 | 1.358E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.952E-06 | 1.132E-03 |
|---|
| nucleus localization | GO:0051647 |  | 7.04E-06 | 2.313E-03 |
|---|
| syncytium formation by plasma membrane fusion | GO:0000768 |  | 1.003E-05 | 2.882E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.003E-05 | 2.562E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.374E-05 | 3.16E-03 |
|---|
| syncytium formation | GO:0006949 |  | 1.374E-05 | 2.873E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.016E-06 | 2.482E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.016E-06 | 1.241E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.829E-06 | 2.304E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.829E-06 | 1.728E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.829E-06 | 1.382E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 4.232E-06 | 1.723E-03 |
|---|
| syncytium formation by plasma membrane fusion | GO:0000768 |  | 6.029E-06 | 2.104E-03 |
|---|
| syncytium formation | GO:0006949 |  | 8.268E-06 | 2.525E-03 |
|---|
| nucleus localization | GO:0051647 |  | 8.268E-06 | 2.244E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.099E-05 | 2.686E-03 |
|---|
| myotube differentiation | GO:0014902 |  | 1.809E-05 | 4.017E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.048E-06 | 2.521E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.048E-06 | 1.26E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.828E-06 | 1.466E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.828E-06 | 1.1E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.917E-06 | 1.404E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.917E-06 | 1.17E-03 |
|---|
| syncytium formation by plasma membrane fusion | GO:0000768 |  | 6.217E-06 | 2.137E-03 |
|---|
| syncytium formation | GO:0006949 |  | 8.525E-06 | 2.564E-03 |
|---|
| nucleus localization | GO:0051647 |  | 8.525E-06 | 2.279E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.133E-05 | 2.727E-03 |
|---|
| myotube differentiation | GO:0014902 |  | 1.865E-05 | 4.079E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 8.49E-07 | 1.953E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 8.49E-07 | 9.763E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.693E-06 | 1.298E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.693E-06 | 9.732E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.952E-06 | 1.358E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.952E-06 | 1.132E-03 |
|---|
| nucleus localization | GO:0051647 |  | 7.04E-06 | 2.313E-03 |
|---|
| syncytium formation by plasma membrane fusion | GO:0000768 |  | 1.003E-05 | 2.882E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.003E-05 | 2.562E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.374E-05 | 3.16E-03 |
|---|
| syncytium formation | GO:0006949 |  | 1.374E-05 | 2.873E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 8.49E-07 | 1.953E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 8.49E-07 | 9.763E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.693E-06 | 1.298E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.693E-06 | 9.732E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.952E-06 | 1.358E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.952E-06 | 1.132E-03 |
|---|
| nucleus localization | GO:0051647 |  | 7.04E-06 | 2.313E-03 |
|---|
| syncytium formation by plasma membrane fusion | GO:0000768 |  | 1.003E-05 | 2.882E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.003E-05 | 2.562E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.374E-05 | 3.16E-03 |
|---|
| syncytium formation | GO:0006949 |  | 1.374E-05 | 2.873E-03 |
|---|


