Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1134
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3335 | 1.495e-03 | 2.600e-05 | 1.618e-01 | 6.287e-09 |
|---|
| IPC | 0.3378 | 3.538e-01 | 1.828e-01 | 9.367e-01 | 6.059e-02 |
|---|
| Loi | 0.4745 | 6.000e-05 | 0.000e+00 | 5.436e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5985 | 1.000e-06 | 0.000e+00 | 1.158e-01 | 0.000e+00 |
|---|
| Wang | 0.2576 | 7.544e-03 | 6.577e-02 | 2.285e-01 | 1.134e-04 |
|---|
Expression data for subnetwork 1134 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
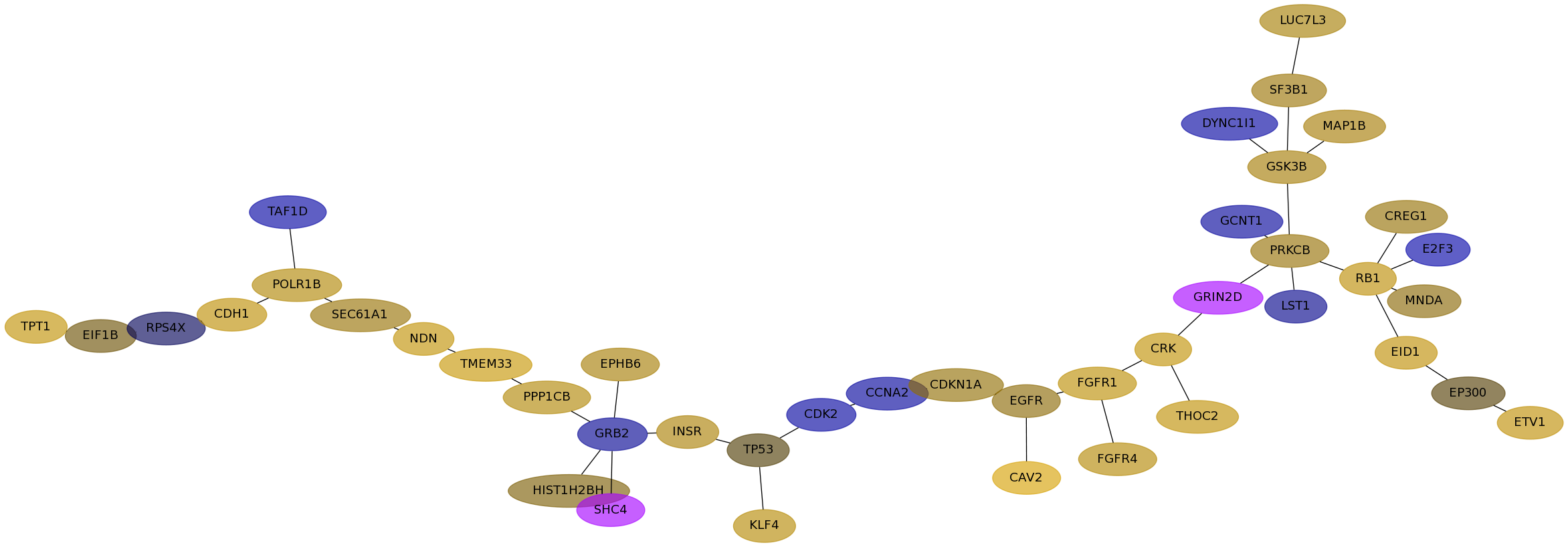
Score for each gene in subnetwork 1134 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| SHC4 |   | 1 | 652 | 1030 | 1037 | undef | -0.088 | 0.034 | undef | undef |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| DYNC1I1 |   | 5 | 148 | 182 | 181 | -0.109 | -0.069 | 0.013 | -0.086 | 0.088 |
|---|
| EIF1B |   | 6 | 120 | 57 | 62 | 0.029 | 0.149 | 0.075 | 0.332 | -0.070 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| POLR1B |   | 11 | 59 | 129 | 126 | 0.157 | 0.062 | -0.036 | -0.079 | 0.139 |
|---|
| HIST1H2BH |   | 1 | 652 | 1030 | 1037 | 0.044 | -0.040 | 0.064 | -0.046 | -0.043 |
|---|
| EPHB6 |   | 7 | 100 | 419 | 407 | 0.122 | 0.008 | 0.042 | -0.219 | -0.059 |
|---|
| ETV1 |   | 11 | 59 | 746 | 724 | 0.214 | 0.046 | 0.054 | -0.065 | 0.141 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| GCNT1 |   | 2 | 391 | 778 | 789 | -0.091 | 0.080 | 0.137 | 0.013 | 0.014 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| EID1 |   | 4 | 198 | 552 | 547 | 0.212 | -0.069 | 0.051 | -0.010 | -0.037 |
|---|
| TAF1D |   | 5 | 148 | 57 | 63 | -0.110 | 0.141 | 0.113 | 0.041 | 0.059 |
|---|
| CREG1 |   | 5 | 148 | 691 | 670 | 0.080 | 0.029 | 0.018 | -0.189 | -0.176 |
|---|
| E2F3 |   | 5 | 148 | 364 | 358 | -0.126 | 0.041 | -0.091 | 0.177 | 0.050 |
|---|
| MNDA |   | 2 | 391 | 1030 | 1032 | 0.060 | 0.174 | -0.210 | -0.004 | -0.148 |
|---|
| LST1 |   | 1 | 652 | 1030 | 1037 | -0.065 | -0.009 | -0.284 | -0.090 | -0.003 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| GRB2 |   | 1 | 652 | 1030 | 1037 | -0.077 | -0.040 | -0.159 | -0.036 | -0.024 |
|---|
| RPS4X |   | 1 | 652 | 1030 | 1037 | -0.019 | -0.014 | 0.081 | 0.183 | -0.068 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| SEC61A1 |   | 5 | 148 | 271 | 267 | 0.085 | -0.161 | 0.128 | -0.096 | 0.070 |
|---|
| PRKCB |   | 8 | 84 | 33 | 35 | 0.081 | -0.065 | 0.001 | -0.148 | -0.035 |
|---|
| THOC2 |   | 11 | 59 | 419 | 406 | 0.217 | -0.094 | 0.119 | 0.068 | 0.072 |
|---|
| KLF4 |   | 3 | 265 | 271 | 280 | 0.174 | 0.035 | 0.060 | -0.026 | -0.025 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| NDN |   | 3 | 265 | 756 | 748 | 0.229 | -0.097 | 0.169 | 0.083 | 0.245 |
|---|
| PPP1CB |   | 1 | 652 | 1030 | 1037 | 0.159 | -0.074 | 0.091 | -0.022 | 0.025 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| TPT1 |   | 1 | 652 | 1030 | 1037 | 0.227 | 0.042 | 0.149 | -0.046 | -0.029 |
|---|
| TMEM33 |   | 3 | 265 | 1030 | 1008 | 0.264 | -0.074 | -0.077 | -0.187 | 0.033 |
|---|
GO Enrichment output for subnetwork 1134 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.58E-06 | 3.634E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.897E-06 | 2.182E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.958E-06 | 1.501E-03 |
|---|
| dendrite development | GO:0016358 |  | 2.41E-06 | 1.386E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.869E-06 | 1.32E-03 |
|---|
| interphase | GO:0051325 |  | 3.283E-06 | 1.259E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.309E-06 | 1.087E-03 |
|---|
| calcium ion transport | GO:0006816 |  | 6.519E-06 | 1.874E-03 |
|---|
| divalent metal ion transport | GO:0070838 |  | 6.91E-06 | 1.766E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.252E-05 | 2.88E-03 |
|---|
| negative regulation of leukocyte proliferation | GO:0070664 |  | 4.189E-05 | 8.758E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.928E-07 | 7.153E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.635E-07 | 5.662E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.738E-07 | 4.672E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.511E-06 | 9.228E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.511E-06 | 7.382E-04 |
|---|
| dendrite development | GO:0016358 |  | 1.56E-06 | 6.352E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 4.446E-06 | 1.552E-03 |
|---|
| negative regulation of leukocyte proliferation | GO:0070664 |  | 1.484E-05 | 4.533E-03 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 2.104E-05 | 5.712E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.153E-05 | 5.261E-03 |
|---|
| response to UV | GO:0009411 |  | 2.44E-05 | 5.418E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.202E-07 | 1.251E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.992E-07 | 7.209E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.382E-07 | 7.525E-04 |
|---|
| interphase | GO:0051325 |  | 1.127E-06 | 6.779E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.149E-06 | 1.034E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.149E-06 | 8.616E-04 |
|---|
| dendrite development | GO:0016358 |  | 2.48E-06 | 8.523E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 7.346E-06 | 2.209E-03 |
|---|
| negative regulation of leukocyte proliferation | GO:0070664 |  | 2.106E-05 | 5.63E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.551E-05 | 6.138E-03 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 2.849E-05 | 6.232E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.58E-06 | 3.634E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.897E-06 | 2.182E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.958E-06 | 1.501E-03 |
|---|
| dendrite development | GO:0016358 |  | 2.41E-06 | 1.386E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.869E-06 | 1.32E-03 |
|---|
| interphase | GO:0051325 |  | 3.283E-06 | 1.259E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.309E-06 | 1.087E-03 |
|---|
| calcium ion transport | GO:0006816 |  | 6.519E-06 | 1.874E-03 |
|---|
| divalent metal ion transport | GO:0070838 |  | 6.91E-06 | 1.766E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.252E-05 | 2.88E-03 |
|---|
| negative regulation of leukocyte proliferation | GO:0070664 |  | 4.189E-05 | 8.758E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.58E-06 | 3.634E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.897E-06 | 2.182E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.958E-06 | 1.501E-03 |
|---|
| dendrite development | GO:0016358 |  | 2.41E-06 | 1.386E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.869E-06 | 1.32E-03 |
|---|
| interphase | GO:0051325 |  | 3.283E-06 | 1.259E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.309E-06 | 1.087E-03 |
|---|
| calcium ion transport | GO:0006816 |  | 6.519E-06 | 1.874E-03 |
|---|
| divalent metal ion transport | GO:0070838 |  | 6.91E-06 | 1.766E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.252E-05 | 2.88E-03 |
|---|
| negative regulation of leukocyte proliferation | GO:0070664 |  | 4.189E-05 | 8.758E-03 |
|---|


