Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1114
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3336 | 1.490e-03 | 2.600e-05 | 1.615e-01 | 6.257e-09 |
|---|
| IPC | 0.3419 | 3.471e-01 | 1.769e-01 | 9.353e-01 | 5.743e-02 |
|---|
| Loi | 0.4733 | 6.400e-05 | 0.000e+00 | 5.501e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5971 | 1.000e-06 | 0.000e+00 | 1.189e-01 | 0.000e+00 |
|---|
| Wang | 0.2570 | 7.748e-03 | 6.688e-02 | 2.314e-01 | 1.199e-04 |
|---|
Expression data for subnetwork 1114 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
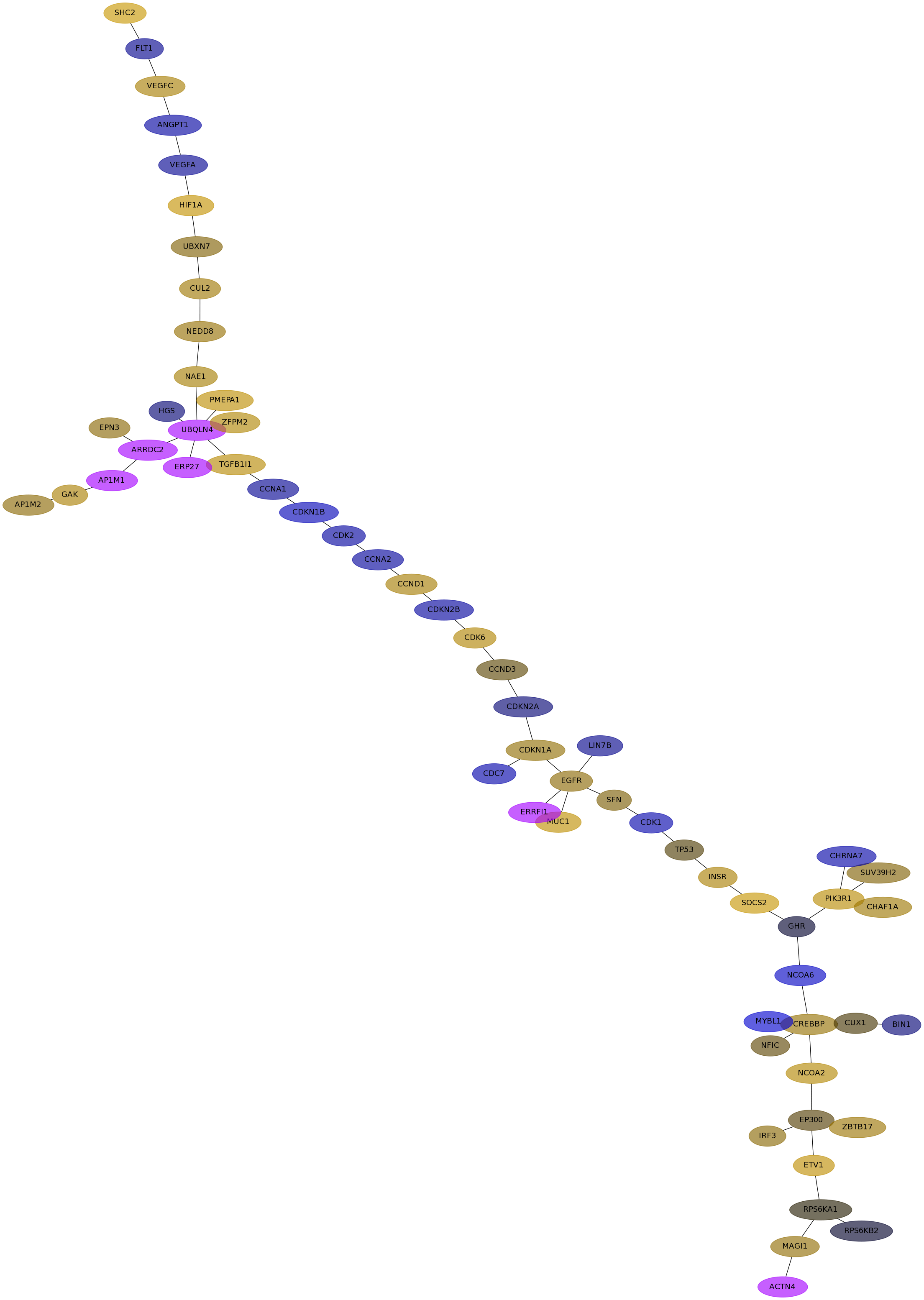
Score for each gene in subnetwork 1114 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| CDKN2B |   | 4 | 198 | 937 | 910 | -0.099 | 0.066 | 0.050 | -0.028 | 0.153 |
|---|
| BIN1 |   | 1 | 652 | 937 | 951 | -0.038 | -0.062 | -0.161 | -0.209 | -0.047 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| RPS6KA1 |   | 8 | 84 | 696 | 669 | 0.005 | -0.208 | 0.084 | -0.322 | -0.071 |
|---|
| SOCS2 |   | 4 | 198 | 859 | 837 | 0.262 | -0.130 | -0.028 | 0.051 | 0.027 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| IRF3 |   | 1 | 652 | 937 | 951 | 0.062 | -0.058 | -0.014 | -0.165 | -0.212 |
|---|
| SHC2 |   | 2 | 391 | 937 | 928 | 0.281 | -0.053 | 0.284 | -0.036 | 0.078 |
|---|
| UBXN7 |   | 6 | 120 | 148 | 156 | 0.049 | -0.055 | 0.153 | 0.224 | 0.175 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| CREBBP |   | 5 | 148 | 444 | 440 | 0.084 | -0.104 | 0.102 | 0.093 | 0.002 |
|---|
| LIN7B |   | 2 | 391 | 206 | 220 | -0.065 | 0.047 | -0.123 | -0.141 | -0.147 |
|---|
| AP1M2 |   | 2 | 391 | 816 | 808 | 0.064 | -0.082 | 0.154 | -0.068 | 0.056 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| SUV39H2 |   | 3 | 265 | 859 | 843 | 0.049 | 0.150 | -0.097 | -0.057 | 0.028 |
|---|
| NCOA6 |   | 1 | 652 | 937 | 951 | -0.239 | -0.179 | -0.123 | -0.167 | 0.133 |
|---|
| CDC7 |   | 1 | 652 | 937 | 951 | -0.134 | -0.005 | -0.168 | 0.025 | 0.126 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| CCND3 |   | 1 | 652 | 937 | 951 | 0.021 | 0.053 | 0.056 | 0.015 | -0.171 |
|---|
| AP1M1 |   | 1 | 652 | 937 | 951 | undef | -0.094 | -0.097 | undef | undef |
|---|
| CDKN1B |   | 2 | 391 | 937 | 928 | -0.180 | 0.120 | -0.171 | 0.088 | -0.005 |
|---|
| ARRDC2 |   | 2 | 391 | 937 | 928 | undef | 0.192 | -0.100 | undef | undef |
|---|
| ERRFI1 |   | 2 | 391 | 937 | 928 | undef | 0.076 | 0.215 | undef | undef |
|---|
| MYBL1 |   | 1 | 652 | 937 | 951 | -0.310 | -0.097 | 0.038 | 0.044 | 0.106 |
|---|
| ETV1 |   | 11 | 59 | 746 | 724 | 0.214 | 0.046 | 0.054 | -0.065 | 0.141 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| NAE1 |   | 1 | 652 | 937 | 951 | 0.123 | 0.012 | -0.007 | -0.189 | 0.199 |
|---|
| NCOA2 |   | 1 | 652 | 937 | 951 | 0.170 | -0.078 | 0.080 | -0.159 | 0.057 |
|---|
| RPS6KB2 |   | 2 | 391 | 937 | 928 | -0.006 | 0.033 | -0.161 | 0.131 | -0.000 |
|---|
| VEGFC |   | 1 | 652 | 937 | 951 | 0.125 | 0.018 | 0.123 | -0.010 | 0.245 |
|---|
| CHRNA7 |   | 1 | 652 | 937 | 951 | -0.126 | 0.101 | -0.014 | 0.086 | -0.022 |
|---|
| ERP27 |   | 1 | 652 | 937 | 951 | undef | -0.244 | 0.258 | undef | undef |
|---|
| ZBTB17 |   | 2 | 391 | 937 | 928 | 0.094 | 0.067 | 0.256 | -0.145 | 0.255 |
|---|
| UBQLN4 |   | 2 | 391 | 937 | 928 | undef | 0.143 | -0.114 | undef | undef |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CUX1 |   | 8 | 84 | 271 | 266 | 0.012 | -0.042 | 0.256 | -0.121 | 0.040 |
|---|
| ZFPM2 |   | 4 | 198 | 148 | 162 | 0.160 | 0.001 | 0.092 | 0.113 | 0.379 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| CHAF1A |   | 2 | 391 | 710 | 709 | 0.102 | -0.006 | -0.147 | 0.022 | -0.057 |
|---|
| EPN3 |   | 2 | 391 | 937 | 928 | 0.061 | 0.185 | 0.179 | 0.001 | 0.081 |
|---|
| NEDD8 |   | 3 | 265 | 536 | 537 | 0.083 | 0.250 | -0.085 | 0.251 | -0.073 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| HGS |   | 1 | 652 | 937 | 951 | -0.037 | -0.015 | -0.078 | -0.037 | 0.020 |
|---|
| PMEPA1 |   | 4 | 198 | 937 | 910 | 0.208 | -0.085 | 0.137 | 0.167 | 0.102 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| CUL2 |   | 3 | 265 | 206 | 208 | 0.103 | 0.124 | -0.192 | -0.090 | 0.166 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| FLT1 |   | 3 | 265 | 364 | 362 | -0.068 | 0.109 | 0.069 | 0.124 | 0.177 |
|---|
| ACTN4 |   | 6 | 120 | 859 | 834 | undef | 0.035 | 0.180 | undef | undef |
|---|
| NFIC |   | 1 | 652 | 937 | 951 | 0.021 | -0.150 | 0.192 | -0.055 | 0.177 |
|---|
| GAK |   | 3 | 265 | 816 | 804 | 0.134 | -0.008 | 0.127 | -0.162 | 0.090 |
|---|
| MAGI1 |   | 2 | 391 | 937 | 928 | 0.073 | 0.094 | 0.020 | 0.073 | 0.004 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| ANGPT1 |   | 2 | 391 | 937 | 928 | -0.105 | 0.106 | -0.167 | 0.084 | 0.250 |
|---|
| CDKN2A |   | 10 | 67 | 364 | 351 | -0.037 | 0.263 | -0.113 | 0.111 | -0.105 |
|---|
| SFN |   | 3 | 265 | 806 | 794 | 0.044 | 0.141 | 0.270 | 0.023 | 0.101 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
GO Enrichment output for subnetwork 1114 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.872E-12 | 4.305E-09 |
|---|
| interphase | GO:0051325 |  | 2.426E-12 | 2.79E-09 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.033E-11 | 7.919E-09 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.769E-11 | 5.617E-08 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.439E-08 | 6.618E-06 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.343E-08 | 1.281E-05 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 6.656E-08 | 2.187E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.632E-07 | 4.693E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.554E-07 | 6.528E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 5.618E-07 | 1.292E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.285E-06 | 2.687E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.106E-13 | 1.98E-09 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.273E-12 | 5.22E-09 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.032E-11 | 2.469E-08 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 9.315E-11 | 5.689E-08 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 4.161E-10 | 2.033E-07 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 9.963E-09 | 4.057E-06 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.661E-08 | 9.288E-06 |
|---|
| lung development | GO:0030324 |  | 2.683E-08 | 8.194E-06 |
|---|
| respiratory tube development | GO:0030323 |  | 3.623E-08 | 9.833E-06 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 4.164E-08 | 1.017E-05 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 4.164E-08 | 9.248E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.123E-12 | 2.701E-09 |
|---|
| interphase | GO:0051325 |  | 1.592E-12 | 1.915E-09 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.508E-12 | 6.021E-09 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.024E-11 | 3.022E-08 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.29E-10 | 6.207E-08 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 5.76E-10 | 2.31E-07 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 9.757E-09 | 3.353E-06 |
|---|
| lung development | GO:0030324 |  | 3.376E-08 | 1.015E-05 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 3.41E-08 | 9.116E-06 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.457E-08 | 8.317E-06 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 4.459E-08 | 9.752E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.872E-12 | 4.305E-09 |
|---|
| interphase | GO:0051325 |  | 2.426E-12 | 2.79E-09 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.033E-11 | 7.919E-09 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.769E-11 | 5.617E-08 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.439E-08 | 6.618E-06 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.343E-08 | 1.281E-05 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 6.656E-08 | 2.187E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.632E-07 | 4.693E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.554E-07 | 6.528E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 5.618E-07 | 1.292E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.285E-06 | 2.687E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.872E-12 | 4.305E-09 |
|---|
| interphase | GO:0051325 |  | 2.426E-12 | 2.79E-09 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.033E-11 | 7.919E-09 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.769E-11 | 5.617E-08 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.439E-08 | 6.618E-06 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.343E-08 | 1.281E-05 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 6.656E-08 | 2.187E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.632E-07 | 4.693E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.554E-07 | 6.528E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 5.618E-07 | 1.292E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.285E-06 | 2.687E-04 |
|---|


