Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1103
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3352 | 1.384e-03 | 2.300e-05 | 1.564e-01 | 4.980e-09 |
|---|
| IPC | 0.3407 | 3.489e-01 | 1.786e-01 | 9.357e-01 | 5.830e-02 |
|---|
| Loi | 0.4726 | 6.600e-05 | 0.000e+00 | 5.536e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5950 | 1.000e-06 | 0.000e+00 | 1.235e-01 | 0.000e+00 |
|---|
| Wang | 0.2562 | 7.989e-03 | 6.817e-02 | 2.346e-01 | 1.278e-04 |
|---|
Expression data for subnetwork 1103 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
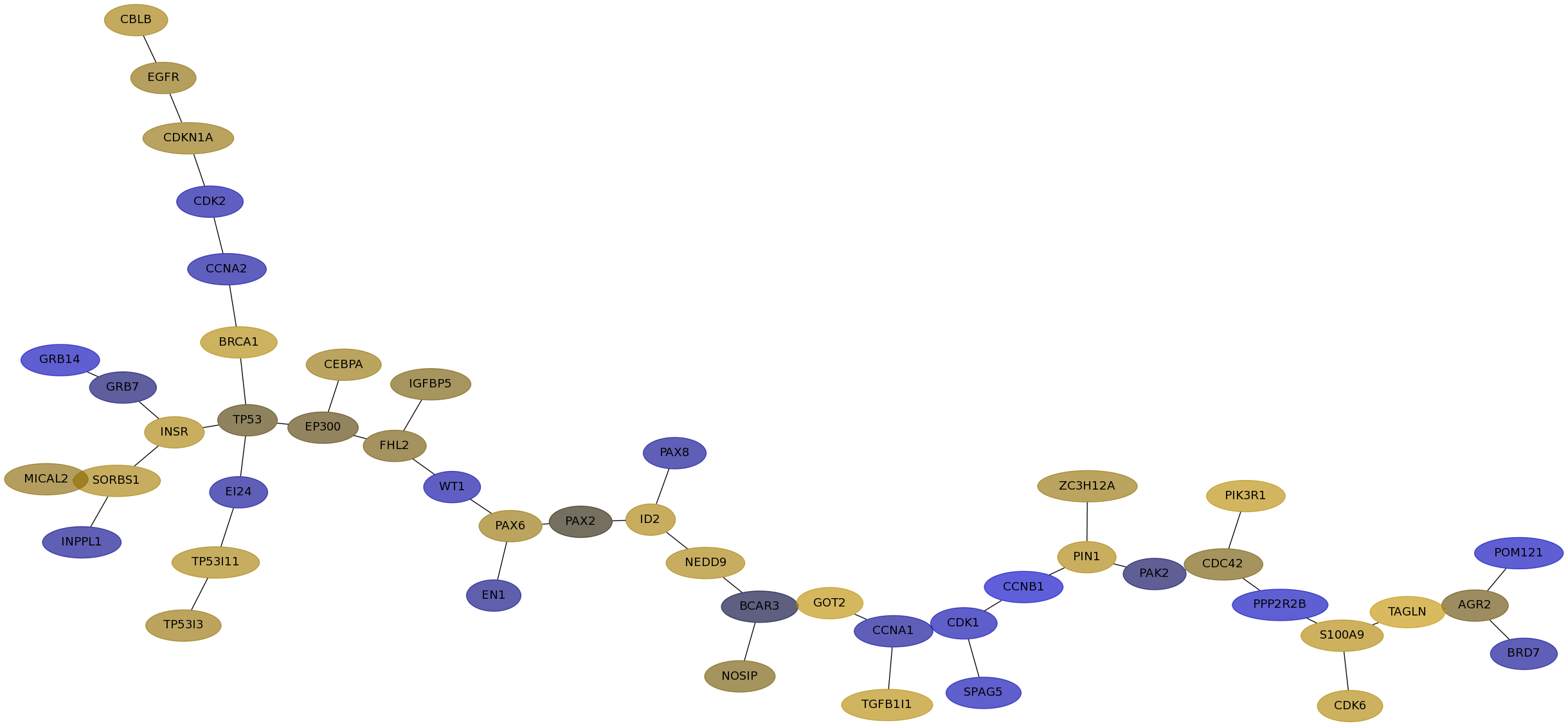
Score for each gene in subnetwork 1103 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| INPPL1 |   | 2 | 391 | 679 | 678 | -0.065 | -0.122 | 0.072 | -0.087 | 0.156 |
|---|
| PAX6 |   | 11 | 59 | 283 | 274 | 0.081 | 0.259 | -0.003 | 0.112 | 0.168 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| GRB14 |   | 4 | 198 | 977 | 965 | -0.189 | 0.113 | -0.129 | 0.143 | 0.138 |
|---|
| NEDD9 |   | 2 | 391 | 1052 | 1044 | 0.126 | 0.020 | 0.023 | 0.032 | 0.222 |
|---|
| PAX2 |   | 1 | 652 | 1052 | 1064 | 0.005 | 0.100 | 0.076 | -0.113 | 0.186 |
|---|
| AGR2 |   | 1 | 652 | 1052 | 1064 | 0.025 | 0.194 | 0.076 | -0.005 | -0.028 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| ID2 |   | 1 | 652 | 1052 | 1064 | 0.129 | 0.142 | -0.087 | -0.020 | -0.194 |
|---|
| CEBPA |   | 3 | 265 | 1052 | 1036 | 0.080 | -0.058 | 0.005 | -0.100 | 0.039 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| TP53I11 |   | 1 | 652 | 1052 | 1064 | 0.122 | 0.022 | 0.058 | -0.285 | 0.119 |
|---|
| NOSIP |   | 1 | 652 | 1052 | 1064 | 0.036 | 0.308 | -0.028 | 0.002 | 0.055 |
|---|
| CCNB1 |   | 2 | 391 | 364 | 375 | -0.278 | 0.241 | -0.056 | 0.001 | -0.043 |
|---|
| SPAG5 |   | 4 | 198 | 492 | 499 | -0.159 | 0.132 | -0.243 | 0.031 | 0.084 |
|---|
| POM121 |   | 5 | 148 | 419 | 415 | -0.195 | -0.197 | 0.101 | 0.079 | 0.017 |
|---|
| EN1 |   | 1 | 652 | 1052 | 1064 | -0.050 | 0.196 | 0.024 | 0.011 | 0.102 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| GOT2 |   | 2 | 391 | 1052 | 1044 | 0.210 | -0.061 | 0.100 | -0.238 | 0.105 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| EI24 |   | 8 | 84 | 339 | 336 | -0.074 | 0.051 | 0.078 | 0.107 | 0.070 |
|---|
| BRD7 |   | 4 | 198 | 906 | 894 | -0.068 | -0.090 | 0.019 | -0.188 | -0.086 |
|---|
| WT1 |   | 8 | 84 | 283 | 278 | -0.106 | 0.254 | -0.039 | 0.036 | 0.172 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| TAGLN |   | 4 | 198 | 823 | 807 | 0.243 | -0.079 | 0.246 | 0.170 | 0.175 |
|---|
| MICAL2 |   | 3 | 265 | 182 | 189 | 0.061 | -0.012 | 0.143 | -0.042 | 0.161 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| FHL2 |   | 5 | 148 | 206 | 202 | 0.033 | 0.143 | 0.239 | -0.033 | 0.294 |
|---|
| CBLB |   | 3 | 265 | 182 | 189 | 0.109 | -0.168 | 0.266 | -0.248 | 0.223 |
|---|
| BRCA1 |   | 5 | 148 | 598 | 575 | 0.157 | -0.201 | -0.078 | -0.087 | 0.168 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| TP53I3 |   | 2 | 391 | 1052 | 1044 | 0.081 | -0.065 | 0.133 | 0.123 | -0.007 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| BCAR3 |   | 2 | 391 | 584 | 578 | -0.009 | 0.271 | 0.123 | 0.259 | 0.066 |
|---|
| ZC3H12A |   | 2 | 391 | 1052 | 1044 | 0.079 | 0.034 | 0.001 | 0.094 | -0.114 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| PAX8 |   | 4 | 198 | 778 | 766 | -0.072 | 0.008 | -0.083 | 0.032 | -0.002 |
|---|
| IGFBP5 |   | 3 | 265 | 444 | 447 | 0.038 | -0.076 | 0.110 | 0.074 | 0.181 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
GO Enrichment output for subnetwork 1103 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.182E-06 | 2.719E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.182E-06 | 1.359E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.559E-06 | 1.195E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.355E-06 | 1.354E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.21E-06 | 1.477E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.908E-06 | 1.498E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.107E-06 | 1.349E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 4.107E-06 | 1.181E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.361E-06 | 1.114E-03 |
|---|
| interphase | GO:0051325 |  | 4.987E-06 | 1.147E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.634E-06 | 1.178E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.581E-07 | 1.608E-03 |
|---|
| nuclear migration | GO:0007097 |  | 9.485E-07 | 1.159E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 9.485E-07 | 7.724E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.703E-06 | 1.04E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.036E-06 | 9.948E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.641E-06 | 1.076E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.641E-06 | 9.219E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.809E-06 | 8.577E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.267E-06 | 8.869E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.952E-06 | 9.654E-04 |
|---|
| myeloid cell differentiation | GO:0030099 |  | 4.877E-06 | 1.083E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.932E-07 | 2.39E-03 |
|---|
| nuclear migration | GO:0007097 |  | 1.223E-06 | 1.471E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.223E-06 | 9.809E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.134E-06 | 1.284E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.134E-06 | 1.027E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.296E-06 | 9.207E-04 |
|---|
| interphase | GO:0051325 |  | 2.754E-06 | 9.465E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.842E-06 | 8.547E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.348E-06 | 8.951E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.405E-06 | 8.192E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.918E-06 | 8.571E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.182E-06 | 2.719E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.182E-06 | 1.359E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.559E-06 | 1.195E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.355E-06 | 1.354E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.21E-06 | 1.477E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.908E-06 | 1.498E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.107E-06 | 1.349E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 4.107E-06 | 1.181E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.361E-06 | 1.114E-03 |
|---|
| interphase | GO:0051325 |  | 4.987E-06 | 1.147E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.634E-06 | 1.178E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.182E-06 | 2.719E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.182E-06 | 1.359E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.559E-06 | 1.195E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.355E-06 | 1.354E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.21E-06 | 1.477E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.908E-06 | 1.498E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.107E-06 | 1.349E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 4.107E-06 | 1.181E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.361E-06 | 1.114E-03 |
|---|
| interphase | GO:0051325 |  | 4.987E-06 | 1.147E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.634E-06 | 1.178E-03 |
|---|


