Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1102
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3354 | 1.372e-03 | 2.300e-05 | 1.559e-01 | 4.919e-09 |
|---|
| IPC | 0.3396 | 3.508e-01 | 1.802e-01 | 9.361e-01 | 5.916e-02 |
|---|
| Loi | 0.4724 | 6.600e-05 | 0.000e+00 | 5.549e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5954 | 1.000e-06 | 0.000e+00 | 1.225e-01 | 0.000e+00 |
|---|
| Wang | 0.2559 | 8.090e-03 | 6.871e-02 | 2.360e-01 | 1.312e-04 |
|---|
Expression data for subnetwork 1102 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
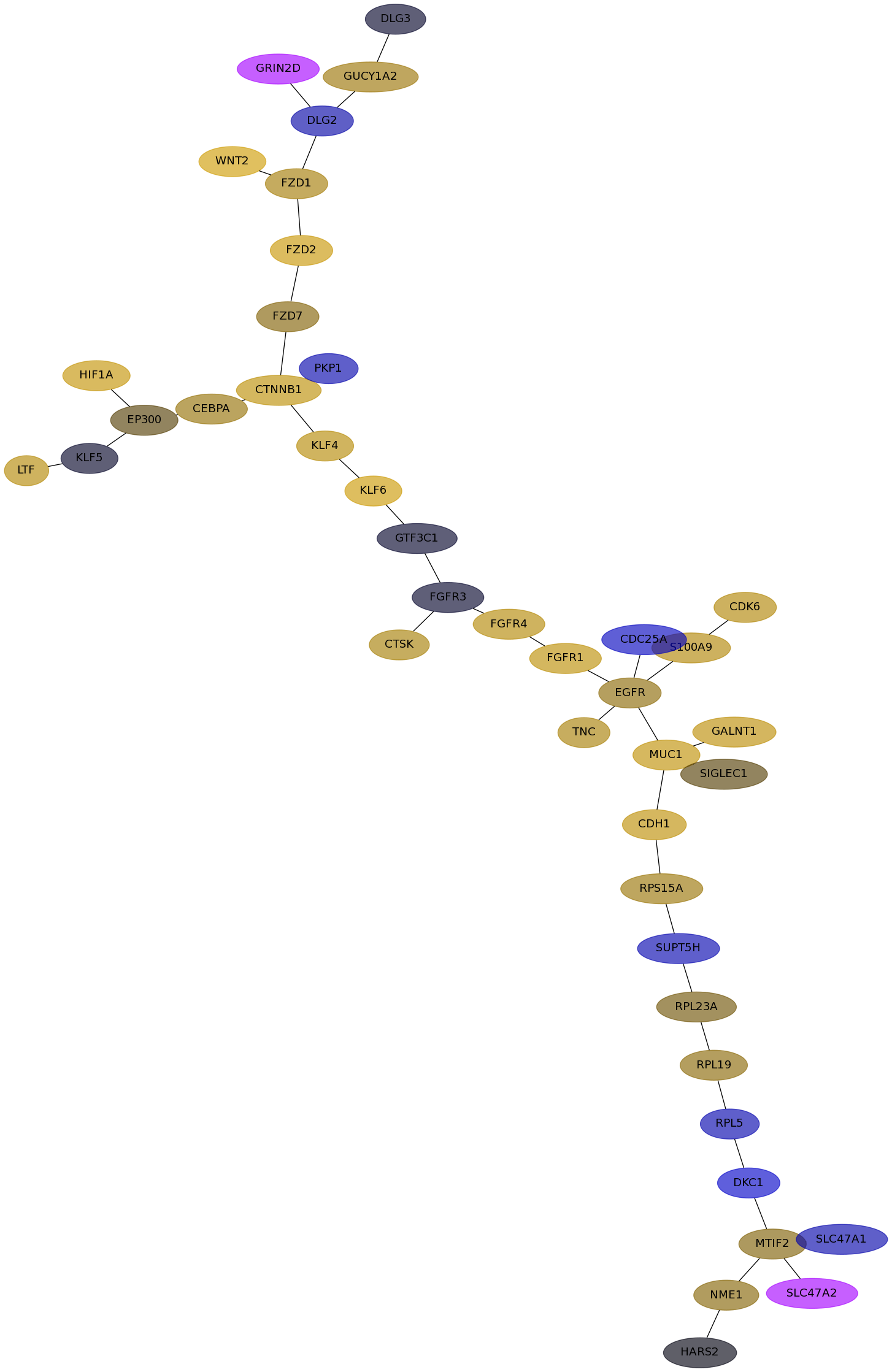
Score for each gene in subnetwork 1102 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| WNT2 |   | 2 | 391 | 647 | 646 | 0.314 | 0.223 | -0.064 | -0.148 | 0.186 |
|---|
| CEBPA |   | 3 | 265 | 1052 | 1036 | 0.080 | -0.058 | 0.005 | -0.100 | 0.039 |
|---|
| FZD2 |   | 8 | 84 | 552 | 540 | 0.273 | -0.067 | -0.081 | -0.067 | 0.081 |
|---|
| RPL5 |   | 4 | 198 | 889 | 873 | -0.145 | -0.000 | 0.128 | 0.220 | -0.110 |
|---|
| TNC |   | 2 | 391 | 339 | 352 | 0.125 | -0.154 | 0.282 | 0.113 | 0.072 |
|---|
| LTF |   | 7 | 100 | 492 | 495 | 0.168 | -0.082 | 0.060 | -0.268 | -0.280 |
|---|
| RPL23A |   | 10 | 67 | 57 | 52 | 0.031 | 0.208 | -0.007 | 0.024 | 0.055 |
|---|
| GALNT1 |   | 3 | 265 | 889 | 874 | 0.200 | 0.133 | -0.061 | -0.181 | 0.234 |
|---|
| FGFR3 |   | 5 | 148 | 963 | 938 | -0.006 | 0.160 | 0.071 | 0.018 | -0.111 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| SLC47A1 |   | 1 | 652 | 1084 | 1091 | -0.134 | 0.131 | -0.105 | -0.234 | 0.145 |
|---|
| RPL19 |   | 1 | 652 | 1084 | 1091 | 0.060 | 0.009 | 0.043 | -0.022 | 0.063 |
|---|
| HARS2 |   | 4 | 198 | 606 | 597 | -0.002 | 0.049 | 0.360 | -0.168 | -0.199 |
|---|
| DLG3 |   | 1 | 652 | 1084 | 1091 | -0.005 | 0.023 | 0.155 | 0.010 | -0.039 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| FZD1 |   | 2 | 391 | 647 | 646 | 0.117 | -0.281 | 0.189 | -0.114 | -0.023 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| CDC25A |   | 5 | 148 | 444 | 440 | -0.220 | 0.113 | -0.170 | 0.151 | -0.003 |
|---|
| GTF3C1 |   | 1 | 652 | 1084 | 1091 | -0.006 | 0.144 | 0.102 | -0.083 | 0.076 |
|---|
| MTIF2 |   | 4 | 198 | 492 | 499 | 0.047 | 0.013 | 0.029 | -0.088 | 0.031 |
|---|
| SUPT5H |   | 9 | 77 | 1 | 18 | -0.150 | -0.085 | 0.007 | -0.298 | -0.068 |
|---|
| DKC1 |   | 1 | 652 | 1084 | 1091 | -0.273 | -0.002 | -0.115 | 0.143 | 0.085 |
|---|
| GUCY1A2 |   | 1 | 652 | 1084 | 1091 | 0.092 | 0.135 | -0.058 | -0.088 | 0.131 |
|---|
| KLF6 |   | 2 | 391 | 1084 | 1072 | 0.295 | 0.173 | 0.001 | 0.099 | 0.115 |
|---|
| CTSK |   | 1 | 652 | 1084 | 1091 | 0.123 | 0.028 | 0.032 | -0.018 | 0.118 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| FZD7 |   | 5 | 148 | 552 | 541 | 0.051 | -0.219 | 0.062 | 0.062 | -0.059 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| RPS15A |   | 1 | 652 | 1084 | 1091 | 0.089 | 0.221 | -0.019 | 0.189 | -0.060 |
|---|
| NME1 |   | 7 | 100 | 492 | 495 | 0.054 | 0.267 | 0.106 | 0.046 | -0.035 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| KLF4 |   | 3 | 265 | 271 | 280 | 0.174 | 0.035 | 0.060 | -0.026 | -0.025 |
|---|
| DLG2 |   | 1 | 652 | 1084 | 1091 | -0.120 | -0.080 | 0.010 | 0.066 | -0.092 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| PKP1 |   | 2 | 391 | 563 | 567 | -0.141 | 0.123 | 0.104 | 0.127 | 0.011 |
|---|
| SLC47A2 |   | 3 | 265 | 492 | 503 | undef | undef | 0.083 | undef | undef |
|---|
| SIGLEC1 |   | 3 | 265 | 963 | 950 | 0.017 | 0.045 | -0.114 | -0.107 | -0.052 |
|---|
GO Enrichment output for subnetwork 1102 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.044E-10 | 7E-07 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 5.414E-08 | 6.226E-05 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 1.59E-07 | 1.219E-04 |
|---|
| cell growth | GO:0016049 |  | 6.97E-07 | 4.008E-04 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 1.216E-06 | 5.596E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.216E-06 | 4.663E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.564E-06 | 5.14E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 1.564E-06 | 4.498E-04 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.11E-06 | 5.392E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 2.729E-06 | 6.277E-04 |
|---|
| UTP metabolic process | GO:0046051 |  | 4.353E-06 | 9.102E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 8.647E-11 | 2.112E-07 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.768E-08 | 3.382E-05 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 5.856E-08 | 4.769E-05 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.113E-07 | 6.796E-05 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.793E-07 | 1.365E-04 |
|---|
| cell growth | GO:0016049 |  | 3.066E-07 | 1.248E-04 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 3.13E-07 | 1.092E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 8.48E-07 | 2.589E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 9.731E-07 | 2.641E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 9.731E-07 | 2.377E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.553E-06 | 3.45E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.127E-10 | 2.712E-07 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.933E-08 | 2.326E-05 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 8.322E-08 | 6.675E-05 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.581E-07 | 9.508E-05 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 3.643E-07 | 1.753E-04 |
|---|
| cell growth | GO:0016049 |  | 4.216E-07 | 1.69E-04 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 4.442E-07 | 1.527E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 8.319E-07 | 2.502E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.269E-06 | 3.392E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 1.269E-06 | 3.053E-04 |
|---|
| UTP metabolic process | GO:0046051 |  | 2.025E-06 | 4.429E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.044E-10 | 7E-07 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 5.414E-08 | 6.226E-05 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 1.59E-07 | 1.219E-04 |
|---|
| cell growth | GO:0016049 |  | 6.97E-07 | 4.008E-04 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 1.216E-06 | 5.596E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.216E-06 | 4.663E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.564E-06 | 5.14E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 1.564E-06 | 4.498E-04 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.11E-06 | 5.392E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 2.729E-06 | 6.277E-04 |
|---|
| UTP metabolic process | GO:0046051 |  | 4.353E-06 | 9.102E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.044E-10 | 7E-07 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 5.414E-08 | 6.226E-05 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 1.59E-07 | 1.219E-04 |
|---|
| cell growth | GO:0016049 |  | 6.97E-07 | 4.008E-04 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 1.216E-06 | 5.596E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.216E-06 | 4.663E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.564E-06 | 5.14E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 1.564E-06 | 4.498E-04 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.11E-06 | 5.392E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 2.729E-06 | 6.277E-04 |
|---|
| UTP metabolic process | GO:0046051 |  | 4.353E-06 | 9.102E-04 |
|---|


