Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1064
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3371 | 1.272e-03 | 2.000e-05 | 1.508e-01 | 3.837e-09 |
|---|
| IPC | 0.3348 | 3.587e-01 | 1.872e-01 | 9.377e-01 | 6.297e-02 |
|---|
| Loi | 0.4677 | 8.200e-05 | 0.000e+00 | 5.792e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5893 | 2.000e-06 | 0.000e+00 | 1.361e-01 | 0.000e+00 |
|---|
| Wang | 0.2559 | 8.087e-03 | 6.869e-02 | 2.360e-01 | 1.311e-04 |
|---|
Expression data for subnetwork 1064 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
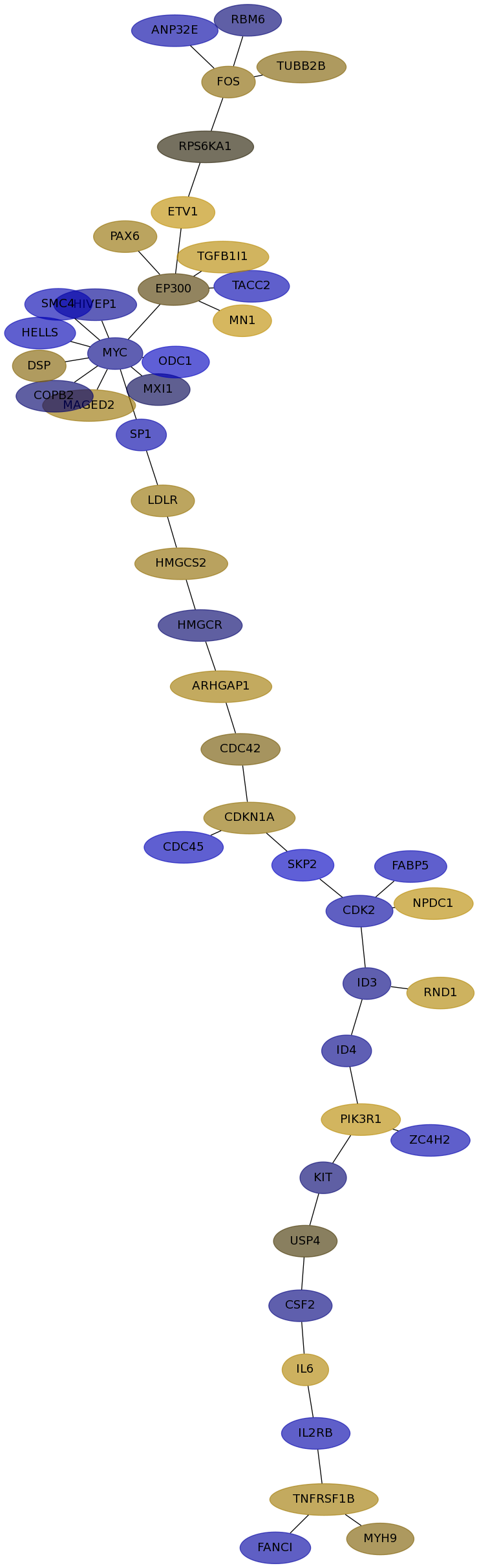
Score for each gene in subnetwork 1064 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| USP4 |   | 6 | 120 | 823 | 803 | 0.012 | -0.093 | 0.147 | 0.097 | 0.054 |
|---|
| MXI1 |   | 1 | 652 | 1319 | 1323 | -0.016 | -0.004 | 0.140 | 0.158 | 0.033 |
|---|
| PAX6 |   | 11 | 59 | 283 | 274 | 0.081 | 0.259 | -0.003 | 0.112 | 0.168 |
|---|
| RPS6KA1 |   | 8 | 84 | 696 | 669 | 0.005 | -0.208 | 0.084 | -0.322 | -0.071 |
|---|
| LDLR |   | 3 | 265 | 679 | 667 | 0.085 | -0.041 | 0.218 | 0.039 | 0.064 |
|---|
| MYH9 |   | 2 | 391 | 1319 | 1320 | 0.047 | -0.103 | 0.227 | 0.007 | 0.165 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| COPB2 |   | 1 | 652 | 1319 | 1323 | -0.031 | 0.087 | 0.184 | -0.204 | 0.080 |
|---|
| FANCI |   | 4 | 198 | 832 | 809 | -0.111 | 0.145 | -0.194 | 0.104 | -0.036 |
|---|
| RND1 |   | 2 | 391 | 444 | 458 | 0.158 | -0.046 | 0.085 | 0.022 | 0.113 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| TACC2 |   | 9 | 77 | 148 | 154 | -0.151 | -0.073 | 0.122 | 0.181 | 0.180 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| RBM6 |   | 10 | 67 | 148 | 152 | -0.036 | -0.082 | 0.265 | -0.004 | 0.111 |
|---|
| ARHGAP1 |   | 3 | 265 | 129 | 139 | 0.108 | -0.167 | 0.297 | 0.001 | 0.251 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| ETV1 |   | 11 | 59 | 746 | 724 | 0.214 | 0.046 | 0.054 | -0.065 | 0.141 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| SMC4 |   | 1 | 652 | 1319 | 1323 | -0.159 | 0.247 | -0.111 | 0.214 | 0.094 |
|---|
| HMGCR |   | 3 | 265 | 129 | 139 | -0.029 | -0.075 | 0.226 | 0.026 | -0.016 |
|---|
| MN1 |   | 6 | 120 | 148 | 156 | 0.213 | -0.166 | -0.047 | 0.150 | 0.011 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| HMGCS2 |   | 3 | 265 | 129 | 139 | 0.073 | -0.277 | 0.226 | -0.194 | -0.085 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| FABP5 |   | 2 | 391 | 665 | 660 | -0.155 | 0.152 | -0.177 | 0.225 | 0.141 |
|---|
| IL2RB |   | 2 | 391 | 715 | 711 | -0.133 | 0.079 | -0.240 | -0.175 | -0.068 |
|---|
| CSF2 |   | 1 | 652 | 1319 | 1323 | -0.047 | -0.028 | -0.076 | -0.133 | -0.197 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| NPDC1 |   | 3 | 265 | 339 | 348 | 0.190 | -0.019 | 0.221 | 0.044 | -0.146 |
|---|
| DSP |   | 7 | 100 | 206 | 198 | 0.052 | 0.054 | 0.235 | -0.119 | 0.053 |
|---|
| ODC1 |   | 5 | 148 | 398 | 387 | -0.220 | -0.006 | -0.099 | 0.209 | -0.107 |
|---|
| TNFRSF1B |   | 12 | 55 | 563 | 544 | 0.109 | 0.024 | -0.187 | -0.096 | -0.106 |
|---|
| MAGED2 |   | 2 | 391 | 1065 | 1059 | 0.089 | -0.104 | 0.198 | 0.022 | -0.020 |
|---|
| CDC45 |   | 4 | 198 | 847 | 830 | -0.177 | 0.134 | -0.171 | 0.185 | -0.085 |
|---|
| ID3 |   | 2 | 391 | 746 | 751 | -0.051 | 0.072 | -0.006 | 0.192 | 0.057 |
|---|
| HELLS |   | 1 | 652 | 1319 | 1323 | -0.171 | -0.050 | -0.018 | 0.199 | 0.107 |
|---|
| IL6 |   | 3 | 265 | 715 | 705 | 0.155 | 0.257 | -0.106 | 0.226 | 0.035 |
|---|
| TUBB2B |   | 5 | 148 | 148 | 161 | 0.048 | -0.036 | 0.304 | 0.045 | 0.139 |
|---|
| ANP32E |   | 6 | 120 | 148 | 156 | -0.114 | 0.166 | -0.139 | 0.191 | 0.118 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| HIVEP1 |   | 6 | 120 | 419 | 410 | -0.072 | -0.199 | 0.069 | 0.000 | -0.042 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
GO Enrichment output for subnetwork 1064 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| myeloid leukocyte differentiation | GO:0002573 |  | 1.989E-08 | 4.575E-05 |
|---|
| establishment of organelle localization | GO:0051656 |  | 6.917E-08 | 7.955E-05 |
|---|
| organelle localization | GO:0051640 |  | 2.022E-07 | 1.55E-04 |
|---|
| regulation of survival gene product expression | GO:0045884 |  | 3.488E-07 | 2.006E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 3.063E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.659E-07 | 2.553E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.328E-06 | 4.363E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.335E-06 | 3.839E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.535E-06 | 3.924E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.317E-06 | 5.329E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.317E-06 | 4.844E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| myeloid leukocyte differentiation | GO:0002573 |  | 6.584E-09 | 1.608E-05 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.844E-09 | 1.08E-05 |
|---|
| establishment of organelle localization | GO:0051656 |  | 1.33E-08 | 1.083E-05 |
|---|
| organelle localization | GO:0051640 |  | 5.292E-08 | 3.232E-05 |
|---|
| regulation of survival gene product expression | GO:0045884 |  | 1.157E-07 | 5.655E-05 |
|---|
| nuclear migration | GO:0007097 |  | 3.796E-07 | 1.546E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 3.796E-07 | 1.325E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.796E-07 | 1.159E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 4.431E-07 | 1.203E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.059E-06 | 2.586E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.059E-06 | 2.351E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| myeloid leukocyte differentiation | GO:0002573 |  | 7.263E-09 | 1.747E-05 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.873E-08 | 2.253E-05 |
|---|
| establishment of organelle localization | GO:0051656 |  | 2.55E-08 | 2.045E-05 |
|---|
| organelle localization | GO:0051640 |  | 1.011E-07 | 6.082E-05 |
|---|
| regulation of survival gene product expression | GO:0045884 |  | 1.801E-07 | 8.666E-05 |
|---|
| nuclear migration | GO:0007097 |  | 5.302E-07 | 2.126E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 5.302E-07 | 1.822E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.302E-07 | 1.595E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 7.622E-07 | 2.038E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.258E-07 | 2.228E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 9.258E-07 | 2.025E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| myeloid leukocyte differentiation | GO:0002573 |  | 1.989E-08 | 4.575E-05 |
|---|
| establishment of organelle localization | GO:0051656 |  | 6.917E-08 | 7.955E-05 |
|---|
| organelle localization | GO:0051640 |  | 2.022E-07 | 1.55E-04 |
|---|
| regulation of survival gene product expression | GO:0045884 |  | 3.488E-07 | 2.006E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 3.063E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.659E-07 | 2.553E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.328E-06 | 4.363E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.335E-06 | 3.839E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.535E-06 | 3.924E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.317E-06 | 5.329E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.317E-06 | 4.844E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| myeloid leukocyte differentiation | GO:0002573 |  | 1.989E-08 | 4.575E-05 |
|---|
| establishment of organelle localization | GO:0051656 |  | 6.917E-08 | 7.955E-05 |
|---|
| organelle localization | GO:0051640 |  | 2.022E-07 | 1.55E-04 |
|---|
| regulation of survival gene product expression | GO:0045884 |  | 3.488E-07 | 2.006E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 3.063E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.659E-07 | 2.553E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.328E-06 | 4.363E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.335E-06 | 3.839E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.535E-06 | 3.924E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.317E-06 | 5.329E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.317E-06 | 4.844E-04 |
|---|


