Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1034
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3384 | 1.196e-03 | 1.800e-05 | 1.468e-01 | 3.161e-09 |
|---|
| IPC | 0.3390 | 3.519e-01 | 1.811e-01 | 9.363e-01 | 5.967e-02 |
|---|
| Loi | 0.4628 | 1.030e-04 | 0.000e+00 | 6.063e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5863 | 2.000e-06 | 0.000e+00 | 1.433e-01 | 0.000e+00 |
|---|
| Wang | 0.2556 | 8.178e-03 | 6.917e-02 | 2.372e-01 | 1.342e-04 |
|---|
Expression data for subnetwork 1034 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
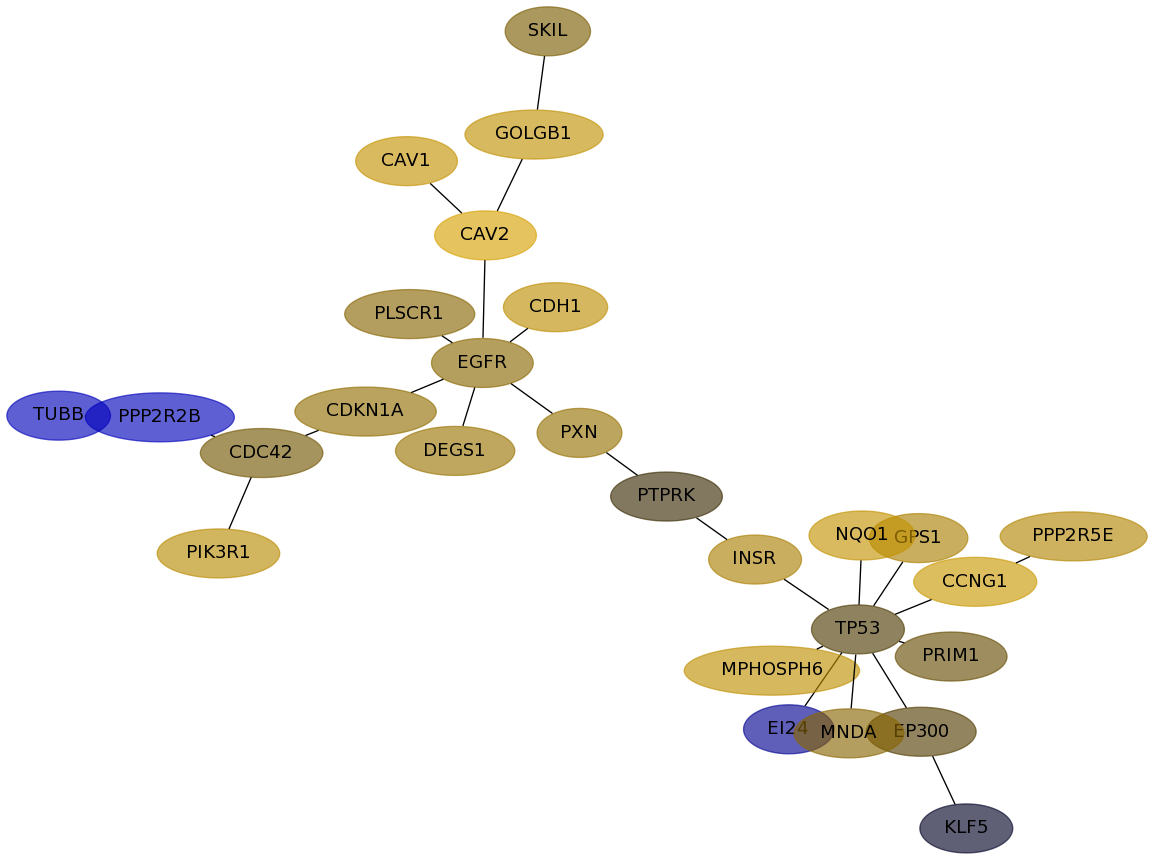
Score for each gene in subnetwork 1034 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| PXN |   | 5 | 148 | 439 | 432 | 0.085 | -0.088 | 0.284 | -0.022 | 0.114 |
|---|
| PLSCR1 |   | 7 | 100 | 634 | 614 | 0.058 | 0.202 | -0.159 | 0.091 | -0.105 |
|---|
| NQO1 |   | 6 | 120 | 536 | 535 | 0.243 | -0.115 | 0.069 | -0.189 | 0.118 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CCNG1 |   | 4 | 198 | 816 | 802 | 0.287 | -0.100 | 0.061 | -0.157 | -0.093 |
|---|
| CAV1 |   | 10 | 67 | 439 | 428 | 0.251 | -0.092 | 0.124 | 0.079 | 0.119 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| MNDA |   | 2 | 391 | 1030 | 1032 | 0.060 | 0.174 | -0.210 | -0.004 | -0.148 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| PRIM1 |   | 3 | 265 | 900 | 892 | 0.027 | 0.153 | -0.219 | 0.059 | 0.091 |
|---|
| SKIL |   | 2 | 391 | 1336 | 1322 | 0.045 | 0.209 | 0.108 | 0.073 | -0.020 |
|---|
| PPP2R5E |   | 3 | 265 | 816 | 804 | 0.160 | 0.001 | 0.191 | 0.018 | 0.094 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| DEGS1 |   | 1 | 652 | 1336 | 1338 | 0.093 | 0.298 | 0.019 | -0.028 | 0.140 |
|---|
| EI24 |   | 8 | 84 | 339 | 336 | -0.074 | 0.051 | 0.078 | 0.107 | 0.070 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| GPS1 |   | 3 | 265 | 900 | 892 | 0.137 | -0.239 | -0.126 | 0.057 | -0.145 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| TUBB |   | 2 | 391 | 398 | 411 | -0.201 | -0.118 | 0.170 | 0.200 | -0.045 |
|---|
| MPHOSPH6 |   | 8 | 84 | 756 | 726 | 0.217 | -0.084 | 0.175 | -0.269 | 0.146 |
|---|
| GOLGB1 |   | 3 | 265 | 117 | 127 | 0.231 | 0.104 | 0.352 | 0.044 | 0.175 |
|---|
| PTPRK |   | 6 | 120 | 148 | 156 | 0.008 | 0.156 | 0.141 | -0.094 | -0.024 |
|---|
GO Enrichment output for subnetwork 1034 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 4.055E-10 | 9.327E-07 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.481E-09 | 4.003E-06 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.013E-08 | 7.763E-06 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 2.64E-08 | 1.518E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 4.686E-08 | 2.156E-05 |
|---|
| cell killing | GO:0001906 |  | 7.953E-08 | 3.049E-05 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 1.878E-07 | 6.17E-05 |
|---|
| nuclear migration | GO:0007097 |  | 2.774E-07 | 7.975E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 2.774E-07 | 7.089E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.774E-07 | 6.38E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 2.774E-07 | 5.8E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 7.724E-11 | 1.887E-07 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.874E-10 | 7.175E-07 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 3.219E-09 | 2.622E-06 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 7.565E-09 | 4.621E-06 |
|---|
| protein oligomerization | GO:0051259 |  | 1.381E-08 | 6.748E-06 |
|---|
| cell killing | GO:0001906 |  | 2.077E-08 | 8.455E-06 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 7.946E-08 | 2.773E-05 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 1.101E-07 | 3.362E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.587E-07 | 4.308E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.587E-07 | 3.877E-05 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 1.9E-07 | 4.219E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 1.186E-10 | 2.854E-07 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 7.628E-10 | 9.176E-07 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 4.939E-09 | 3.961E-06 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 1.16E-08 | 6.979E-06 |
|---|
| protein oligomerization | GO:0051259 |  | 2.39E-08 | 1.15E-05 |
|---|
| cell killing | GO:0001906 |  | 3.183E-08 | 1.277E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.097E-07 | 3.772E-05 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 1.382E-07 | 4.155E-05 |
|---|
| nuclear migration | GO:0007097 |  | 2.191E-07 | 5.857E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.191E-07 | 5.272E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 2.191E-07 | 4.792E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 4.055E-10 | 9.327E-07 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.481E-09 | 4.003E-06 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.013E-08 | 7.763E-06 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 2.64E-08 | 1.518E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 4.686E-08 | 2.156E-05 |
|---|
| cell killing | GO:0001906 |  | 7.953E-08 | 3.049E-05 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 1.878E-07 | 6.17E-05 |
|---|
| nuclear migration | GO:0007097 |  | 2.774E-07 | 7.975E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 2.774E-07 | 7.089E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.774E-07 | 6.38E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 2.774E-07 | 5.8E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 4.055E-10 | 9.327E-07 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.481E-09 | 4.003E-06 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.013E-08 | 7.763E-06 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 2.64E-08 | 1.518E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 4.686E-08 | 2.156E-05 |
|---|
| cell killing | GO:0001906 |  | 7.953E-08 | 3.049E-05 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 1.878E-07 | 6.17E-05 |
|---|
| nuclear migration | GO:0007097 |  | 2.774E-07 | 7.975E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 2.774E-07 | 7.089E-05 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.774E-07 | 6.38E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 2.774E-07 | 5.8E-05 |
|---|


