Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1007
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3395 | 1.139e-03 | 1.700e-05 | 1.437e-01 | 2.783e-09 |
|---|
| IPC | 0.3449 | 3.420e-01 | 1.725e-01 | 9.342e-01 | 5.513e-02 |
|---|
| Loi | 0.4587 | 1.230e-04 | 0.000e+00 | 6.291e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5836 | 2.000e-06 | 0.000e+00 | 1.500e-01 | 0.000e+00 |
|---|
| Wang | 0.2540 | 8.722e-03 | 7.201e-02 | 2.443e-01 | 1.534e-04 |
|---|
Expression data for subnetwork 1007 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
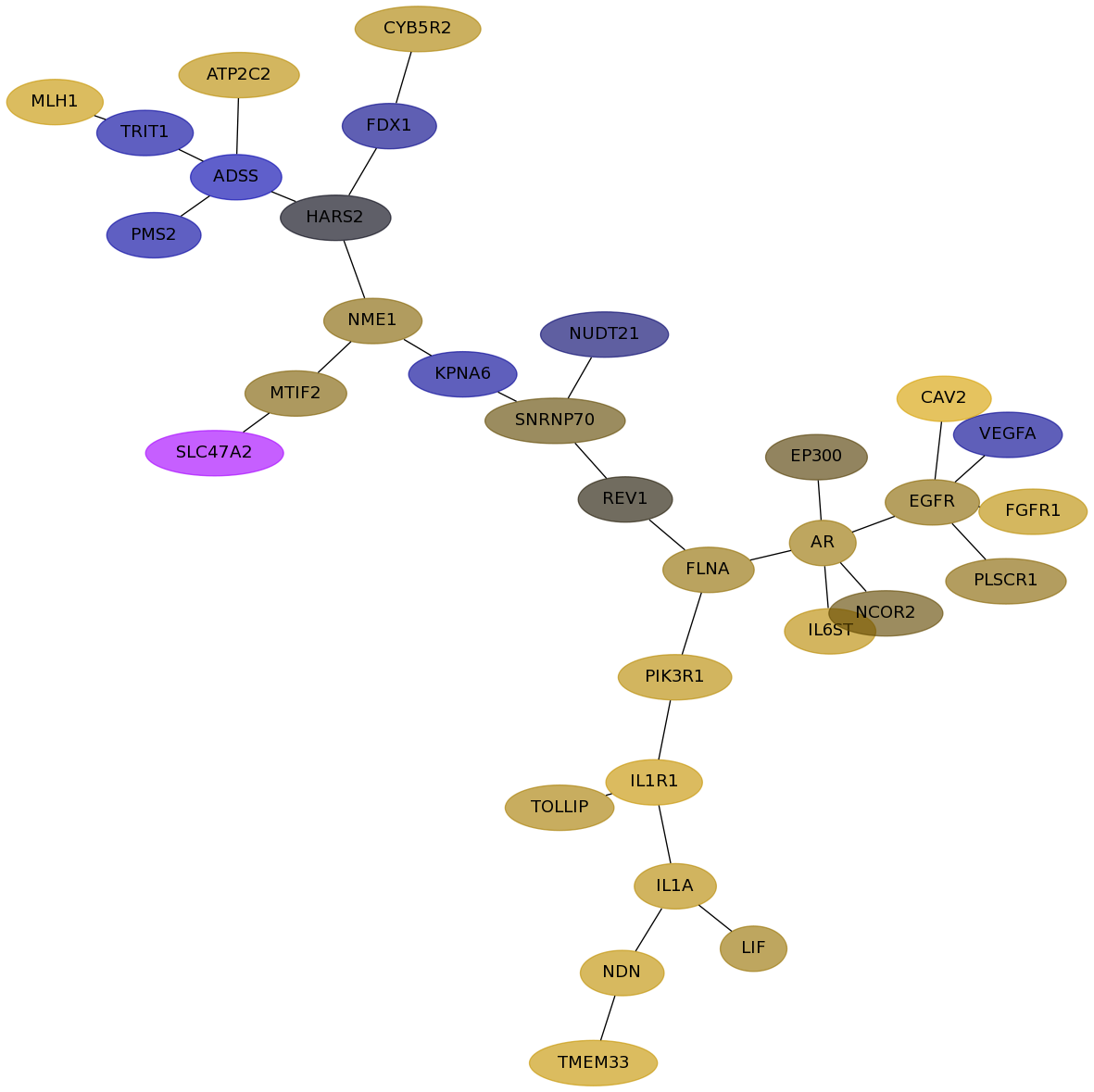
Score for each gene in subnetwork 1007 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| LIF |   | 1 | 652 | 1394 | 1395 | 0.089 | -0.007 | 0.040 | 0.207 | -0.070 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| NCOR2 |   | 11 | 59 | 283 | 274 | 0.024 | -0.096 | 0.211 | -0.163 | 0.114 |
|---|
| PLSCR1 |   | 7 | 100 | 634 | 614 | 0.058 | 0.202 | -0.159 | 0.091 | -0.105 |
|---|
| TRIT1 |   | 1 | 652 | 1394 | 1395 | -0.098 | 0.079 | 0.084 | -0.012 | -0.024 |
|---|
| CYB5R2 |   | 2 | 391 | 584 | 578 | 0.147 | 0.164 | 0.230 | 0.006 | 0.016 |
|---|
| MTIF2 |   | 4 | 198 | 492 | 499 | 0.047 | 0.013 | 0.029 | -0.088 | 0.031 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| ATP2C2 |   | 1 | 652 | 1394 | 1395 | 0.197 | 0.047 | 0.102 | -0.097 | -0.029 |
|---|
| PMS2 |   | 1 | 652 | 1394 | 1395 | -0.104 | 0.029 | 0.095 | -0.017 | -0.026 |
|---|
| MLH1 |   | 1 | 652 | 1394 | 1395 | 0.268 | -0.237 | 0.122 | 0.032 | -0.035 |
|---|
| FLNA |   | 7 | 100 | 206 | 198 | 0.077 | 0.081 | 0.107 | 0.148 | 0.126 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| IL1A |   | 1 | 652 | 1394 | 1395 | 0.180 | -0.177 | 0.023 | 0.086 | undef |
|---|
| TOLLIP |   | 1 | 652 | 1394 | 1395 | 0.129 | 0.033 | 0.003 | -0.205 | -0.025 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| FDX1 |   | 1 | 652 | 1394 | 1395 | -0.059 | 0.101 | -0.097 | 0.044 | -0.054 |
|---|
| IL1R1 |   | 8 | 84 | 536 | 515 | 0.257 | -0.211 | 0.178 | -0.071 | 0.003 |
|---|
| ADSS |   | 1 | 652 | 1394 | 1395 | -0.147 | -0.005 | 0.020 | -0.130 | -0.072 |
|---|
| NUDT21 |   | 1 | 652 | 1394 | 1395 | -0.029 | 0.030 | -0.191 | -0.051 | 0.047 |
|---|
| NME1 |   | 7 | 100 | 492 | 495 | 0.054 | 0.267 | 0.106 | 0.046 | -0.035 |
|---|
| IL6ST |   | 1 | 652 | 1394 | 1395 | 0.196 | -0.038 | 0.078 | 0.028 | -0.145 |
|---|
| NDN |   | 3 | 265 | 756 | 748 | 0.229 | -0.097 | 0.169 | 0.083 | 0.245 |
|---|
| AR |   | 9 | 77 | 283 | 277 | 0.089 | -0.163 | 0.081 | -0.035 | -0.030 |
|---|
| HARS2 |   | 4 | 198 | 606 | 597 | -0.002 | 0.049 | 0.360 | -0.168 | -0.199 |
|---|
| SNRNP70 |   | 3 | 265 | 994 | 971 | 0.024 | -0.129 | 0.130 | -0.059 | 0.036 |
|---|
| SLC47A2 |   | 3 | 265 | 492 | 503 | undef | undef | 0.083 | undef | undef |
|---|
| REV1 |   | 1 | 652 | 1394 | 1395 | 0.004 | 0.090 | -0.027 | 0.165 | 0.039 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| KPNA6 |   | 5 | 148 | 182 | 181 | -0.082 | -0.144 | 0.128 | -0.005 | -0.108 |
|---|
| TMEM33 |   | 3 | 265 | 1030 | 1008 | 0.264 | -0.074 | -0.077 | -0.187 | 0.033 |
|---|
GO Enrichment output for subnetwork 1007 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| leukemia inhibitory factor signaling pathway | GO:0048861 |  | 8.143E-10 | 1.873E-06 |
|---|
| positive regulation vascular endothelial growth factor production | GO:0010575 |  | 3.374E-08 | 3.88E-05 |
|---|
| regulation of vascular endothelial growth factor production | GO:0010574 |  | 5.287E-08 | 4.053E-05 |
|---|
| regulation of JAK-STAT cascade | GO:0046425 |  | 4.297E-07 | 2.471E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 6.074E-07 | 2.794E-04 |
|---|
| positive regulation of muscle hypertrophy | GO:0014742 |  | 9.286E-07 | 3.56E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 9.286E-07 | 3.051E-04 |
|---|
| positive regulation of tyrosine phosphorylation of Stat3 protein | GO:0042517 |  | 1.373E-06 | 3.946E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 1.545E-06 | 3.947E-04 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 1.621E-06 | 3.728E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.621E-06 | 3.389E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| leukemia inhibitory factor signaling pathway | GO:0048861 |  | 1.918E-10 | 4.685E-07 |
|---|
| positive regulation vascular endothelial growth factor production | GO:0010575 |  | 7.977E-09 | 9.743E-06 |
|---|
| regulation of vascular endothelial growth factor production | GO:0010574 |  | 1.251E-08 | 1.019E-05 |
|---|
| regulation of JAK-STAT cascade | GO:0046425 |  | 1.461E-07 | 8.926E-05 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 1.564E-07 | 7.642E-05 |
|---|
| positive regulation of muscle hypertrophy | GO:0014742 |  | 3.122E-07 | 1.271E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 3.928E-07 | 1.371E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 4.948E-07 | 1.511E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 5.454E-07 | 1.481E-04 |
|---|
| positive regulation of tyrosine phosphorylation of Stat3 protein | GO:0042517 |  | 5.505E-07 | 1.345E-04 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 8.711E-07 | 1.935E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| leukemia inhibitory factor signaling pathway | GO:0048861 |  | 3.394E-10 | 8.167E-07 |
|---|
| positive regulation vascular endothelial growth factor production | GO:0010575 |  | 1.41E-08 | 1.696E-05 |
|---|
| regulation of vascular endothelial growth factor production | GO:0010574 |  | 2.21E-08 | 1.773E-05 |
|---|
| regulation of JAK-STAT cascade | GO:0046425 |  | 2.115E-07 | 1.272E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.4E-07 | 1.155E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 4.781E-07 | 1.917E-04 |
|---|
| positive regulation of muscle hypertrophy | GO:0014742 |  | 4.79E-07 | 1.647E-04 |
|---|
| positive regulation of tyrosine phosphorylation of Stat3 protein | GO:0042517 |  | 8.213E-07 | 2.47E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 8.366E-07 | 2.236E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 8.393E-07 | 2.019E-04 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 1.336E-06 | 2.921E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| leukemia inhibitory factor signaling pathway | GO:0048861 |  | 8.143E-10 | 1.873E-06 |
|---|
| positive regulation vascular endothelial growth factor production | GO:0010575 |  | 3.374E-08 | 3.88E-05 |
|---|
| regulation of vascular endothelial growth factor production | GO:0010574 |  | 5.287E-08 | 4.053E-05 |
|---|
| regulation of JAK-STAT cascade | GO:0046425 |  | 4.297E-07 | 2.471E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 6.074E-07 | 2.794E-04 |
|---|
| positive regulation of muscle hypertrophy | GO:0014742 |  | 9.286E-07 | 3.56E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 9.286E-07 | 3.051E-04 |
|---|
| positive regulation of tyrosine phosphorylation of Stat3 protein | GO:0042517 |  | 1.373E-06 | 3.946E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 1.545E-06 | 3.947E-04 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 1.621E-06 | 3.728E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.621E-06 | 3.389E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| leukemia inhibitory factor signaling pathway | GO:0048861 |  | 8.143E-10 | 1.873E-06 |
|---|
| positive regulation vascular endothelial growth factor production | GO:0010575 |  | 3.374E-08 | 3.88E-05 |
|---|
| regulation of vascular endothelial growth factor production | GO:0010574 |  | 5.287E-08 | 4.053E-05 |
|---|
| regulation of JAK-STAT cascade | GO:0046425 |  | 4.297E-07 | 2.471E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 6.074E-07 | 2.794E-04 |
|---|
| positive regulation of muscle hypertrophy | GO:0014742 |  | 9.286E-07 | 3.56E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 9.286E-07 | 3.051E-04 |
|---|
| positive regulation of tyrosine phosphorylation of Stat3 protein | GO:0042517 |  | 1.373E-06 | 3.946E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 1.545E-06 | 3.947E-04 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 1.621E-06 | 3.728E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.621E-06 | 3.389E-04 |
|---|


