Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2592 | 2.513e-02 | 2.795e-03 | 4.780e-01 | 3.358e-05 |
|---|
| IPC | 0.5122 | 1.063e-01 | 2.442e-02 | 8.774e-01 | 2.277e-03 |
|---|
| Loi | 0.3496 | 7.313e-03 | 7.000e-06 | 1.631e-01 | 8.350e-09 |
|---|
| Schmidt | 0.5695 | 4.000e-06 | 0.000e+00 | 1.875e-01 | 0.000e+00 |
|---|
| Wang | 0.1695 | 1.115e-01 | 3.364e-01 | 6.753e-01 | 2.534e-02 |
|---|
Expression data for subnetwork 10 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
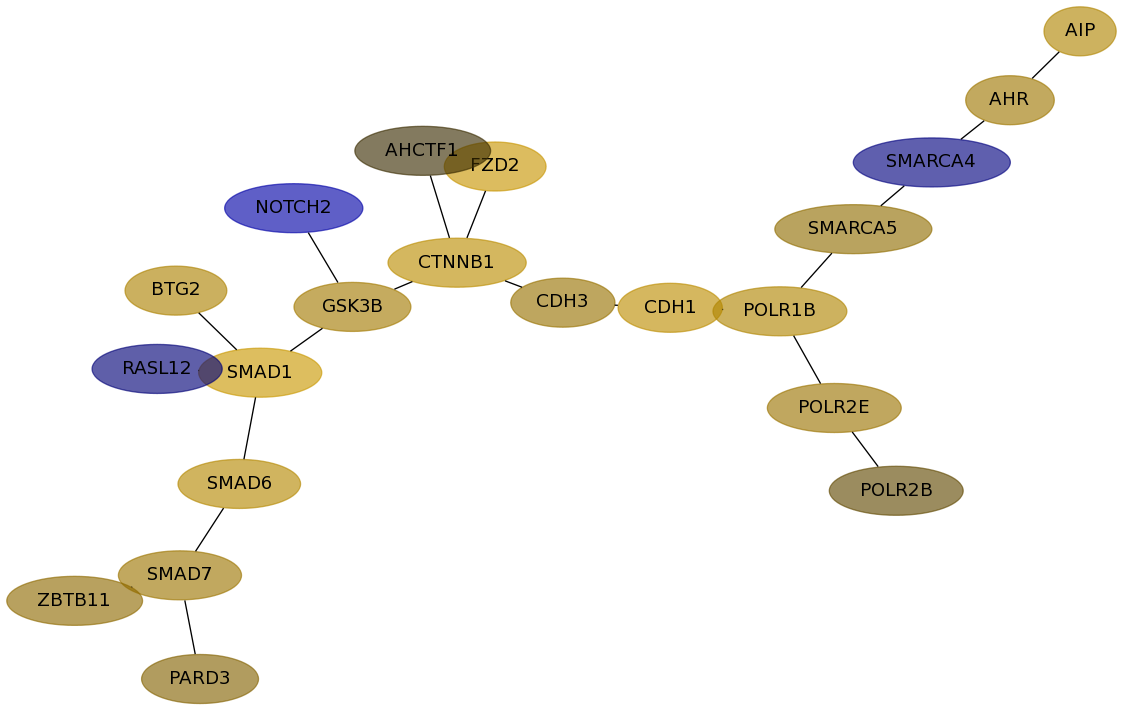
Score for each gene in subnetwork 10 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| BTG2 |   | 6 | 120 | 1 | 21 | 0.146 | -0.029 | -0.093 | -0.111 | -0.170 |
|---|
| SMAD7 |   | 3 | 265 | 665 | 651 | 0.099 | -0.035 | 0.131 | 0.169 | 0.147 |
|---|
| SMARCA4 |   | 4 | 198 | 283 | 289 | -0.050 | 0.080 | 0.202 | -0.071 | -0.116 |
|---|
| NOTCH2 |   | 1 | 652 | 1475 | 1475 | -0.126 | -0.012 | -0.040 | 0.038 | 0.013 |
|---|
| FZD2 |   | 8 | 84 | 552 | 540 | 0.273 | -0.067 | -0.081 | -0.067 | 0.081 |
|---|
| AIP |   | 2 | 391 | 1188 | 1174 | 0.158 | 0.207 | -0.175 | 0.088 | -0.116 |
|---|
| AHCTF1 |   | 1 | 652 | 1475 | 1475 | 0.009 | 0.033 | -0.050 | 0.171 | -0.079 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| POLR2E |   | 4 | 198 | 598 | 577 | 0.095 | 0.127 | 0.039 | 0.166 | -0.018 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| CDH3 |   | 10 | 67 | 1 | 16 | 0.088 | 0.028 | 0.169 | -0.055 | -0.027 |
|---|
| POLR1B |   | 11 | 59 | 129 | 126 | 0.157 | 0.062 | -0.036 | -0.079 | 0.139 |
|---|
| SMARCA5 |   | 1 | 652 | 1475 | 1475 | 0.075 | 0.035 | 0.066 | 0.020 | -0.005 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| ZBTB11 |   | 1 | 652 | 1475 | 1475 | 0.071 | 0.040 | 0.046 | 0.018 | 0.239 |
|---|
| SMAD6 |   | 4 | 198 | 665 | 650 | 0.173 | -0.241 | 0.053 | 0.266 | 0.070 |
|---|
| AHR |   | 1 | 652 | 1475 | 1475 | 0.103 | 0.180 | 0.067 | 0.091 | -0.129 |
|---|
| POLR2B |   | 2 | 391 | 756 | 756 | 0.024 | -0.064 | 0.012 | 0.095 | -0.072 |
|---|
| PARD3 |   | 1 | 652 | 1475 | 1475 | 0.055 | 0.030 | -0.006 | -0.052 | -0.090 |
|---|
| RASL12 |   | 3 | 265 | 665 | 651 | -0.041 | -0.154 | 0.210 | 0.152 | 0.004 |
|---|
GO Enrichment output for subnetwork 10 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| BMP signaling pathway | GO:0030509 |  | 4.165E-07 | 9.58E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.704E-07 | 6.56E-04 |
|---|
| transforming growth factor beta receptor signaling pathway | GO:0007179 |  | 1.478E-06 | 1.133E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 6.831E-06 | 3.928E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 9.704E-06 | 4.464E-03 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 1.744E-05 | 6.685E-03 |
|---|
| cell fate determination | GO:0001709 |  | 1.763E-05 | 5.791E-03 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 2.576E-05 | 7.405E-03 |
|---|
| transcription initiation | GO:0006352 |  | 3.76E-05 | 9.608E-03 |
|---|
| hindbrain development | GO:0030902 |  | 3.997E-05 | 9.194E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 4.12E-05 | 8.615E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.557E-09 | 3.804E-06 |
|---|
| BMP signaling pathway | GO:0030509 |  | 1.657E-07 | 2.024E-04 |
|---|
| transforming growth factor beta receptor signaling pathway | GO:0007179 |  | 5.617E-07 | 4.574E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.489E-06 | 9.092E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.71E-06 | 1.813E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 5.001E-06 | 2.036E-03 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 6.906E-06 | 2.41E-03 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.307E-05 | 3.992E-03 |
|---|
| cell fate determination | GO:0001709 |  | 1.307E-05 | 3.548E-03 |
|---|
| transcription initiation | GO:0006352 |  | 1.484E-05 | 3.626E-03 |
|---|
| hindbrain development | GO:0030902 |  | 1.593E-05 | 3.539E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.401E-09 | 3.37E-06 |
|---|
| BMP signaling pathway | GO:0030509 |  | 2.335E-07 | 2.809E-04 |
|---|
| transforming growth factor beta receptor signaling pathway | GO:0007179 |  | 7.907E-07 | 6.341E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.932E-06 | 1.162E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.811E-06 | 2.315E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 6.484E-06 | 2.6E-03 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 9.212E-06 | 3.166E-03 |
|---|
| cell fate determination | GO:0001709 |  | 1.526E-05 | 4.59E-03 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 1.694E-05 | 4.528E-03 |
|---|
| transcription initiation | GO:0006352 |  | 1.836E-05 | 4.417E-03 |
|---|
| hindbrain development | GO:0030902 |  | 2.064E-05 | 4.515E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| BMP signaling pathway | GO:0030509 |  | 4.165E-07 | 9.58E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.704E-07 | 6.56E-04 |
|---|
| transforming growth factor beta receptor signaling pathway | GO:0007179 |  | 1.478E-06 | 1.133E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 6.831E-06 | 3.928E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 9.704E-06 | 4.464E-03 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 1.744E-05 | 6.685E-03 |
|---|
| cell fate determination | GO:0001709 |  | 1.763E-05 | 5.791E-03 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 2.576E-05 | 7.405E-03 |
|---|
| transcription initiation | GO:0006352 |  | 3.76E-05 | 9.608E-03 |
|---|
| hindbrain development | GO:0030902 |  | 3.997E-05 | 9.194E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 4.12E-05 | 8.615E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| BMP signaling pathway | GO:0030509 |  | 4.165E-07 | 9.58E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.704E-07 | 6.56E-04 |
|---|
| transforming growth factor beta receptor signaling pathway | GO:0007179 |  | 1.478E-06 | 1.133E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 6.831E-06 | 3.928E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 9.704E-06 | 4.464E-03 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 1.744E-05 | 6.685E-03 |
|---|
| cell fate determination | GO:0001709 |  | 1.763E-05 | 5.791E-03 |
|---|
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | GO:0007205 |  | 2.576E-05 | 7.405E-03 |
|---|
| transcription initiation | GO:0006352 |  | 3.76E-05 | 9.608E-03 |
|---|
| hindbrain development | GO:0030902 |  | 3.997E-05 | 9.194E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 4.12E-05 | 8.615E-03 |
|---|


