Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6934
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7488 | 3.849e-03 | 6.430e-04 | 7.831e-01 | 1.938e-06 |
|---|
| Loi | 0.2307 | 7.973e-02 | 1.122e-02 | 4.269e-01 | 3.818e-04 |
|---|
| Schmidt | 0.6764 | 0.000e+00 | 0.000e+00 | 4.183e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7912 | 0.000e+00 | 0.000e+00 | 3.418e-03 | 0.000e+00 |
|---|
| Wang | 0.2640 | 2.372e-03 | 4.094e-02 | 3.826e-01 | 3.716e-05 |
|---|
Expression data for subnetwork 6934 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
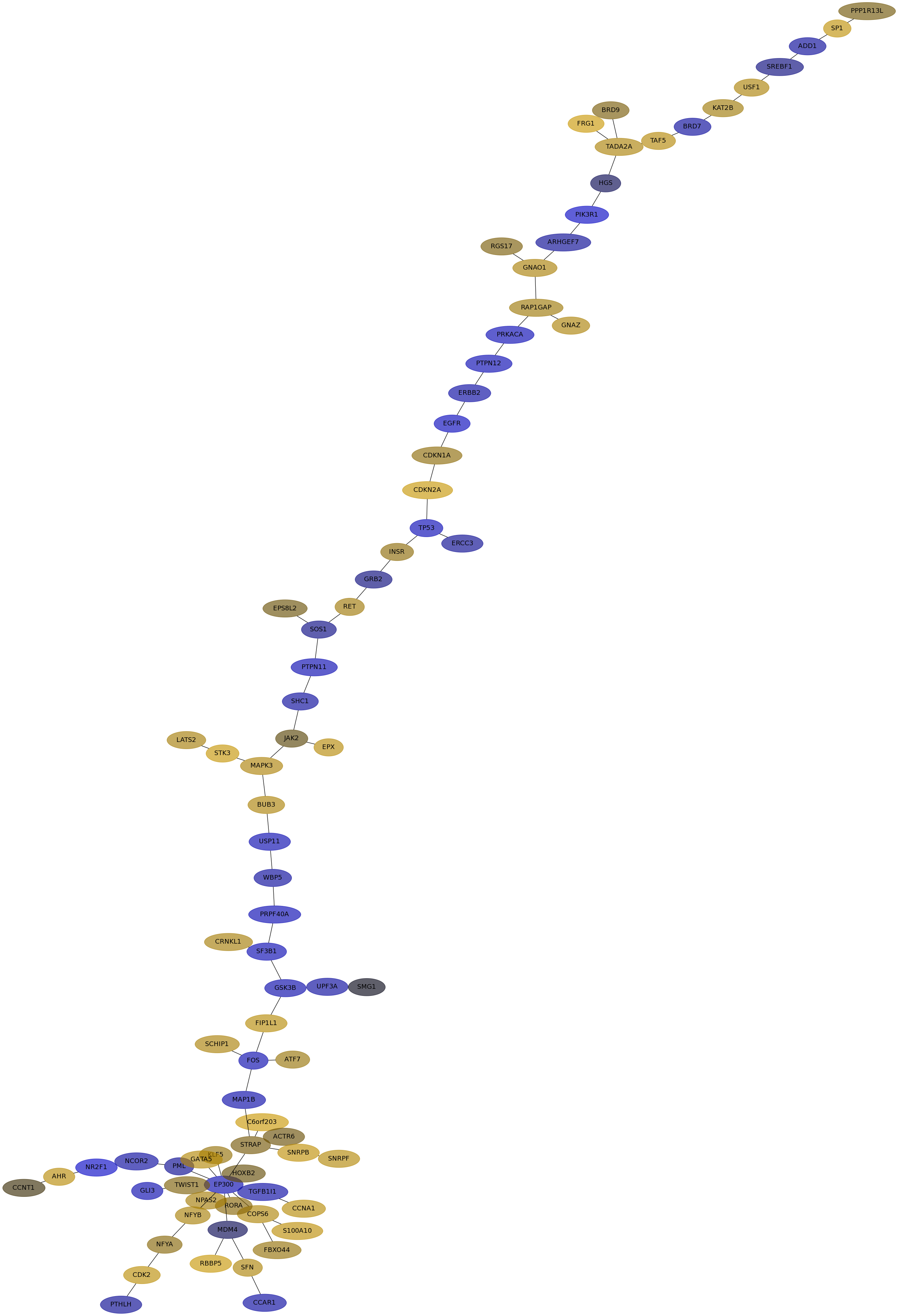
Score for each gene in subnetwork 6934 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ADD1 |   | 8 | 222 | 366 | 373 | -0.085 | 0.116 | 0.035 | undef | -0.098 |
|---|
| PTHLH |   | 2 | 743 | 478 | 490 | -0.070 | 0.111 | 0.047 | -0.033 | -0.091 |
|---|
| NPAS2 |   | 3 | 557 | 366 | 389 | 0.116 | 0.253 | -0.157 | -0.062 | -0.093 |
|---|
| ERCC3 |   | 2 | 743 | 478 | 490 | -0.070 | 0.018 | -0.075 | -0.212 | 0.152 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| RET |   | 7 | 256 | 366 | 376 | 0.098 | 0.063 | -0.179 | undef | -0.159 |
|---|
| MDM4 |   | 9 | 196 | 141 | 147 | -0.016 | 0.004 | 0.164 | 0.107 | 0.105 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| SNRPF |   | 2 | 743 | 478 | 490 | 0.163 | -0.066 | 0.064 | -0.116 | -0.103 |
|---|
| EPX |   | 3 | 557 | 478 | 487 | 0.172 | -0.224 | -0.005 | 0.173 | -0.064 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKACA |   | 2 | 743 | 478 | 490 | -0.163 | 0.181 | 0.104 | -0.038 | -0.144 |
|---|
| RORA |   | 2 | 743 | 478 | 490 | 0.056 | 0.272 | 0.051 | undef | 0.062 |
|---|
| EPS8L2 |   | 2 | 743 | 478 | 490 | 0.028 | 0.287 | -0.160 | undef | 0.106 |
|---|
| USF1 |   | 2 | 743 | 478 | 490 | 0.135 | 0.268 | undef | -0.019 | undef |
|---|
| SHC1 |   | 4 | 440 | 478 | 479 | -0.090 | -0.041 | -0.005 | 0.169 | -0.138 |
|---|
| S100A10 |   | 1 | 1195 | 478 | 523 | 0.201 | 0.168 | 0.068 | 0.050 | 0.213 |
|---|
| GNAZ |   | 15 | 111 | 478 | 457 | 0.134 | -0.116 | 0.308 | 0.267 | 0.011 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| STRAP |   | 7 | 256 | 412 | 414 | 0.035 | -0.128 | 0.074 | 0.154 | -0.049 |
|---|
| TADA2A |   | 4 | 440 | 478 | 479 | 0.135 | -0.054 | 0.204 | -0.115 | -0.127 |
|---|
| CCAR1 |   | 1 | 1195 | 478 | 523 | -0.101 | 0.090 | undef | undef | undef |
|---|
| UPF3A |   | 1 | 1195 | 478 | 523 | -0.092 | -0.014 | 0.065 | -0.082 | 0.031 |
|---|
| ACTR6 |   | 1 | 1195 | 478 | 523 | 0.027 | -0.107 | 0.159 | undef | 0.001 |
|---|
| TAF5 |   | 5 | 360 | 478 | 476 | 0.166 | -0.131 | 0.125 | undef | 0.076 |
|---|
| SNRPB |   | 2 | 743 | 478 | 490 | 0.212 | 0.123 | -0.006 | -0.136 | 0.091 |
|---|
| CRNKL1 |   | 2 | 743 | 478 | 490 | 0.115 | -0.056 | -0.222 | 0.124 | 0.084 |
|---|
| NFYA |   | 1 | 1195 | 478 | 523 | 0.054 | -0.057 | 0.033 | undef | -0.026 |
|---|
| C6orf203 |   | 1 | 1195 | 478 | 523 | 0.285 | -0.074 | undef | 0.050 | undef |
|---|
| CCNT1 |   | 6 | 301 | 478 | 473 | 0.008 | -0.028 | 0.162 | 0.007 | -0.213 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| FIP1L1 |   | 1 | 1195 | 478 | 523 | 0.173 | -0.002 | 0.204 | undef | -0.116 |
|---|
| USP11 |   | 7 | 256 | 478 | 462 | -0.144 | 0.106 | 0.121 | 0.218 | -0.044 |
|---|
| GATA5 |   | 8 | 222 | 366 | 373 | 0.161 | -0.031 | undef | undef | undef |
|---|
| BRD9 |   | 1 | 1195 | 478 | 523 | 0.039 | 0.024 | -0.006 | undef | -0.083 |
|---|
| RAP1GAP |   | 1 | 1195 | 478 | 523 | 0.097 | 0.167 | -0.218 | 0.128 | 0.143 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| SMG1 |   | 1 | 1195 | 478 | 523 | -0.002 | -0.067 | 0.154 | undef | -0.088 |
|---|
| LATS2 |   | 1 | 1195 | 478 | 523 | 0.113 | 0.039 | undef | undef | undef |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| RBBP5 |   | 3 | 557 | 478 | 487 | 0.264 | 0.053 | 0.086 | undef | 0.033 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| RGS17 |   | 2 | 743 | 478 | 490 | 0.040 | 0.066 | -0.090 | 0.302 | 0.011 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| STK3 |   | 4 | 440 | 141 | 165 | 0.254 | 0.007 | 0.106 | 0.326 | 0.198 |
|---|
| SCHIP1 |   | 10 | 167 | 478 | 459 | 0.130 | 0.136 | 0.105 | -0.022 | 0.198 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| SREBF1 |   | 9 | 196 | 478 | 460 | -0.042 | 0.232 | -0.195 | 0.140 | -0.074 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| JAK2 |   | 2 | 743 | 478 | 490 | 0.018 | -0.177 | -0.202 | -0.145 | -0.156 |
|---|
| NFYB |   | 1 | 1195 | 478 | 523 | 0.124 | -0.162 | 0.079 | 0.097 | 0.089 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PRPF40A |   | 2 | 743 | 478 | 490 | -0.153 | -0.047 | -0.133 | undef | 0.155 |
|---|
| WBP5 |   | 1 | 1195 | 478 | 523 | -0.089 | 0.073 | 0.201 | 0.130 | 0.142 |
|---|
| BUB3 |   | 5 | 360 | 478 | 476 | 0.133 | -0.053 | 0.165 | 0.195 | 0.181 |
|---|
| SOS1 |   | 5 | 360 | 478 | 476 | -0.049 | 0.002 | -0.007 | 0.160 | -0.042 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| PTPN12 |   | 6 | 301 | 478 | 473 | -0.159 | 0.153 | 0.031 | 0.083 | 0.042 |
|---|
| FRG1 |   | 1 | 1195 | 478 | 523 | 0.283 | -0.029 | 0.122 | -0.086 | 0.133 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAPK3 |   | 6 | 301 | 141 | 152 | 0.137 | -0.102 | -0.126 | undef | -0.086 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| HGS |   | 1 | 1195 | 478 | 523 | -0.015 | -0.078 | -0.037 | 0.101 | 0.020 |
|---|
| GNAO1 |   | 3 | 557 | 478 | 487 | 0.129 | 0.025 | -0.095 | undef | 0.173 |
|---|
| PML |   | 6 | 301 | 478 | 473 | -0.068 | -0.010 | -0.007 | -0.153 | -0.070 |
|---|
| FBXO44 |   | 1 | 1195 | 478 | 523 | 0.077 | 0.037 | undef | undef | undef |
|---|
| PPP1R13L |   | 2 | 743 | 478 | 490 | 0.032 | 0.351 | -0.104 | 0.042 | 0.173 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| AHR |   | 8 | 222 | 478 | 461 | 0.180 | 0.067 | 0.091 | -0.182 | -0.129 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6934 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.764E-10 | 4.308E-07 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 5.804E-10 | 7.09E-07 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 1.048E-09 | 8.531E-07 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.979E-09 | 1.819E-06 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 3.307E-08 | 1.616E-05 |
|---|
| organ growth | GO:0035265 |  | 4.837E-08 | 1.969E-05 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 4.837E-08 | 1.688E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.233E-08 | 1.903E-05 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 7.142E-08 | 1.939E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 9.627E-08 | 2.352E-05 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.13E-07 | 2.509E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.426E-10 | 5.837E-07 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 7.708E-10 | 9.273E-07 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 1.44E-09 | 1.155E-06 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 3.703E-09 | 2.228E-06 |
|---|
| organ growth | GO:0035265 |  | 6.246E-08 | 3.006E-05 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 6.246E-08 | 2.505E-05 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 7.15E-08 | 2.458E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.556E-08 | 2.874E-05 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 1.237E-07 | 3.307E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.243E-07 | 2.99E-05 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.373E-07 | 3.003E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.578E-10 | 3.63E-07 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 1.455E-09 | 1.673E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.532E-09 | 1.175E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.742E-08 | 1.001E-05 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.067E-07 | 4.91E-05 |
|---|
| organ growth | GO:0035265 |  | 1.204E-07 | 4.617E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.204E-07 | 3.957E-05 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 1.245E-07 | 3.579E-05 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 2.231E-07 | 5.701E-05 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.492E-07 | 5.732E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.675E-07 | 5.594E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell cycle checkpoint | GO:0000075 |  | 4.524E-07 | 8.339E-04 |
|---|
| regulation of mitotic cell cycle | GO:0007346 |  | 6.107E-07 | 5.627E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.345E-06 | 8.264E-04 |
|---|
| response to UV | GO:0009411 |  | 1.902E-06 | 8.762E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 4.566E-06 | 1.683E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 6.676E-06 | 2.051E-03 |
|---|
| ER overload response | GO:0006983 |  | 7.557E-06 | 1.99E-03 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 7.557E-06 | 1.741E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 7.593E-06 | 1.555E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 7.593E-06 | 1.399E-03 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 7.593E-06 | 1.272E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.578E-10 | 3.63E-07 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 1.455E-09 | 1.673E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.532E-09 | 1.175E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.742E-08 | 1.001E-05 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.067E-07 | 4.91E-05 |
|---|
| organ growth | GO:0035265 |  | 1.204E-07 | 4.617E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.204E-07 | 3.957E-05 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 1.245E-07 | 3.579E-05 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 2.231E-07 | 5.701E-05 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.492E-07 | 5.732E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.675E-07 | 5.594E-05 |
|---|



