Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6932
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7487 | 3.850e-03 | 6.430e-04 | 7.832e-01 | 1.939e-06 |
|---|
| Loi | 0.2306 | 7.975e-02 | 1.122e-02 | 4.270e-01 | 3.822e-04 |
|---|
| Schmidt | 0.6764 | 0.000e+00 | 0.000e+00 | 4.180e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.422e-03 | 0.000e+00 |
|---|
| Wang | 0.2640 | 2.370e-03 | 4.093e-02 | 3.826e-01 | 3.711e-05 |
|---|
Expression data for subnetwork 6932 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
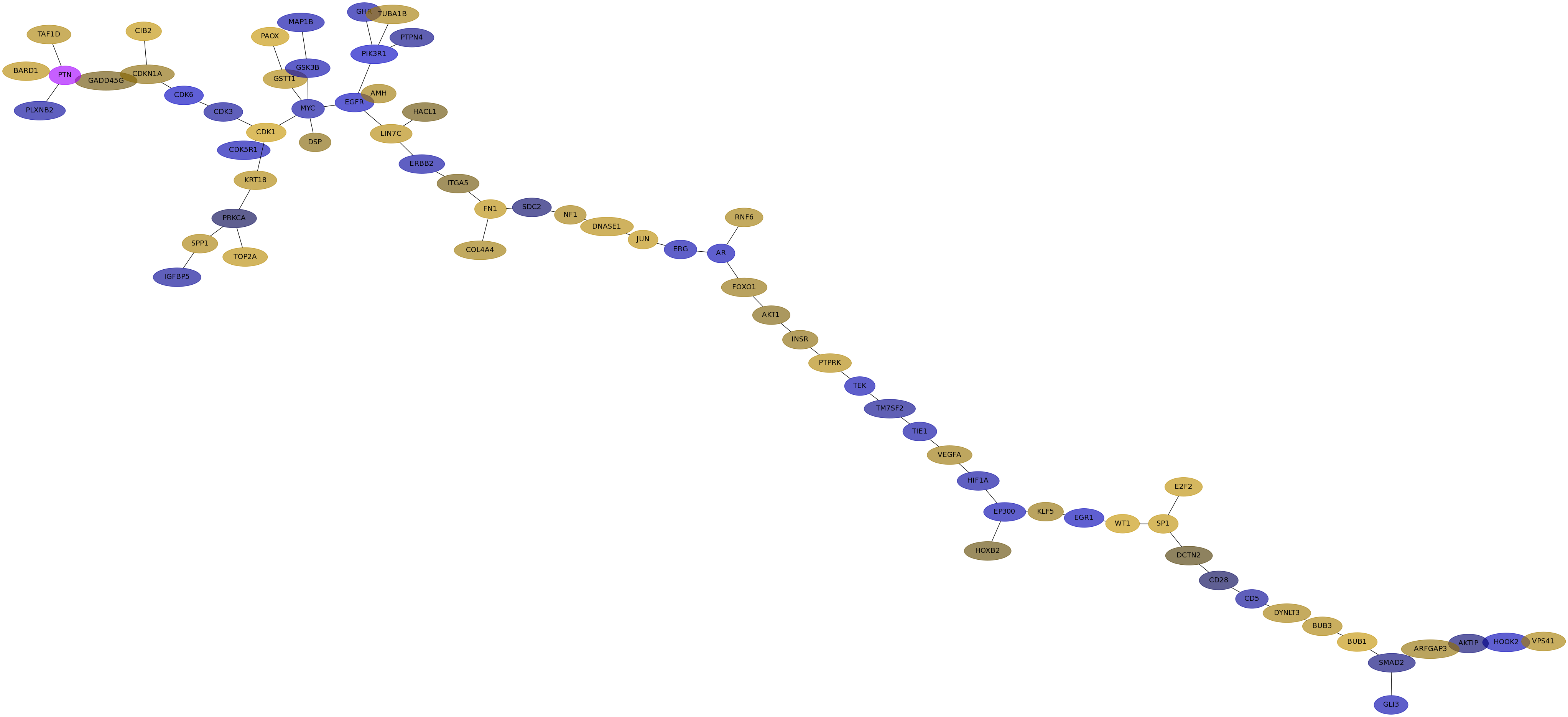
Score for each gene in subnetwork 6932 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| HACL1 |   | 4 | 440 | 590 | 585 | 0.028 | -0.071 | undef | -0.278 | undef |
|---|
| NF1 |   | 9 | 196 | 552 | 541 | 0.110 | -0.052 | -0.032 | 0.090 | -0.094 |
|---|
| DYNLT3 |   | 2 | 743 | 590 | 616 | 0.117 | 0.208 | 0.030 | 0.137 | 0.187 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| HOOK2 |   | 1 | 1195 | 590 | 638 | -0.181 | 0.256 | 0.095 | 0.116 | 0.181 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| CDK3 |   | 7 | 256 | 590 | 570 | -0.076 | 0.189 | -0.041 | -0.089 | 0.082 |
|---|
| CD5 |   | 1 | 1195 | 590 | 638 | -0.080 | -0.131 | -0.112 | -0.221 | 0.030 |
|---|
| AKTIP |   | 1 | 1195 | 590 | 638 | -0.033 | 0.031 | -0.125 | undef | 0.248 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
| DNASE1 |   | 4 | 440 | 590 | 585 | 0.165 | -0.301 | -0.088 | -0.078 | 0.114 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| DSP |   | 7 | 256 | 590 | 570 | 0.054 | 0.235 | -0.119 | -0.033 | 0.053 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| TUBA1B |   | 6 | 301 | 590 | 582 | 0.116 | 0.094 | 0.249 | 0.101 | 0.128 |
|---|
| RNF6 |   | 4 | 440 | 590 | 585 | 0.109 | -0.128 | 0.002 | 0.041 | -0.018 |
|---|
| PAOX |   | 3 | 557 | 1 | 63 | 0.255 | -0.190 | 0.163 | undef | -0.086 |
|---|
| SMAD2 |   | 4 | 440 | 590 | 585 | -0.039 | -0.199 | 0.076 | -0.003 | 0.247 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| FN1 |   | 6 | 301 | 590 | 582 | 0.190 | 0.133 | 0.091 | undef | 0.083 |
|---|
| FOXO1 |   | 3 | 557 | 590 | 598 | 0.072 | 0.094 | -0.021 | -0.147 | 0.221 |
|---|
| DCTN2 |   | 8 | 222 | 366 | 373 | 0.014 | 0.150 | 0.061 | 0.113 | 0.303 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| SDC2 |   | 4 | 440 | 590 | 585 | -0.027 | 0.068 | 0.033 | undef | 0.215 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| AMH |   | 10 | 167 | 366 | 362 | 0.103 | 0.056 | 0.113 | -0.187 | 0.301 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| BUB1 |   | 6 | 301 | 590 | 582 | 0.246 | -0.172 | 0.013 | -0.007 | 0.170 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| ERG |   | 9 | 196 | 179 | 195 | -0.134 | -0.026 | 0.235 | -0.027 | 0.182 |
|---|
| BUB3 |   | 5 | 360 | 478 | 476 | 0.133 | -0.053 | 0.165 | 0.195 | 0.181 |
|---|
| TAF1D |   | 10 | 167 | 590 | 566 | 0.141 | 0.113 | 0.041 | undef | 0.059 |
|---|
| COL4A4 |   | 6 | 301 | 83 | 105 | 0.102 | 0.155 | -0.039 | 0.295 | 0.068 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| TM7SF2 |   | 2 | 743 | 590 | 616 | -0.064 | -0.016 | 0.022 | 0.106 | -0.048 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| VPS41 |   | 1 | 1195 | 590 | 638 | 0.125 | 0.027 | -0.127 | 0.176 | 0.168 |
|---|
| LIN7C |   | 10 | 167 | 590 | 566 | 0.166 | 0.088 | 0.118 | -0.018 | 0.094 |
|---|
| PLXNB2 |   | 4 | 440 | 590 | 585 | -0.092 | 0.308 | -0.136 | undef | -0.062 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| ARFGAP3 |   | 1 | 1195 | 590 | 638 | 0.079 | 0.307 | -0.202 | -0.058 | 0.009 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| PTPN4 |   | 2 | 743 | 318 | 346 | -0.055 | -0.110 | -0.066 | 0.013 | 0.083 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| KRT18 |   | 8 | 222 | 590 | 569 | 0.148 | 0.100 | -0.083 | 0.024 | -0.053 |
|---|
| CD28 |   | 1 | 1195 | 590 | 638 | -0.018 | -0.128 | 0.005 | -0.063 | 0.074 |
|---|
GO Enrichment output for subnetwork 6932 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.582E-08 | 1.119E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.214E-08 | 1.003E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 8.214E-08 | 6.689E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.308E-07 | 7.991E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.008E-07 | 9.812E-05 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 2.077E-07 | 8.455E-05 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 2.134E-07 | 7.448E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.454E-07 | 7.495E-05 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 2.458E-07 | 6.673E-05 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 3.188E-07 | 7.789E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 3.188E-07 | 7.081E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.23E-08 | 1.74E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.296E-07 | 1.559E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.296E-07 | 1.039E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.296E-07 | 1.381E-04 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 3.363E-07 | 1.618E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.521E-07 | 1.412E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.521E-07 | 1.21E-04 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 3.74E-07 | 1.125E-04 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 4.443E-07 | 1.188E-04 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 5.022E-07 | 1.208E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 5.022E-07 | 1.098E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.394E-07 | 5.507E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.394E-07 | 2.754E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.394E-07 | 1.836E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.957E-07 | 4.575E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.988E-07 | 4.595E-04 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 1.108E-06 | 4.249E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.108E-06 | 3.642E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.241E-06 | 3.567E-04 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 1.653E-06 | 4.223E-04 |
|---|
| regulation of angiogenesis | GO:0045765 |  | 2.108E-06 | 4.849E-04 |
|---|
| positive regulation of catabolic process | GO:0009896 |  | 2.252E-06 | 4.71E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell migration | GO:0030334 |  | 1.41E-09 | 2.599E-06 |
|---|
| regulation of locomotion | GO:0040012 |  | 3.732E-09 | 3.439E-06 |
|---|
| regulation of cell motion | GO:0051270 |  | 3.732E-09 | 2.293E-06 |
|---|
| regulation of angiogenesis | GO:0045765 |  | 1.246E-07 | 5.743E-05 |
|---|
| regulation of anatomical structure morphogenesis | GO:0022603 |  | 1.501E-07 | 5.531E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.867E-07 | 2.416E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.182E-06 | 3.113E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 2.435E-06 | 5.609E-04 |
|---|
| heart development | GO:0007507 |  | 2.605E-06 | 5.334E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 3.576E-06 | 6.59E-04 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 3.576E-06 | 5.991E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.394E-07 | 5.507E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.394E-07 | 2.754E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.394E-07 | 1.836E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.957E-07 | 4.575E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.988E-07 | 4.595E-04 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 1.108E-06 | 4.249E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.108E-06 | 3.642E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.241E-06 | 3.567E-04 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 1.653E-06 | 4.223E-04 |
|---|
| regulation of angiogenesis | GO:0045765 |  | 2.108E-06 | 4.849E-04 |
|---|
| positive regulation of catabolic process | GO:0009896 |  | 2.252E-06 | 4.71E-04 |
|---|



