Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6896
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7490 | 3.836e-03 | 6.400e-04 | 7.827e-01 | 1.921e-06 |
|---|
| Loi | 0.2307 | 7.970e-02 | 1.121e-02 | 4.268e-01 | 3.813e-04 |
|---|
| Schmidt | 0.6761 | 0.000e+00 | 0.000e+00 | 4.208e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.428e-03 | 0.000e+00 |
|---|
| Wang | 0.2636 | 2.417e-03 | 4.138e-02 | 3.848e-01 | 3.848e-05 |
|---|
Expression data for subnetwork 6896 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
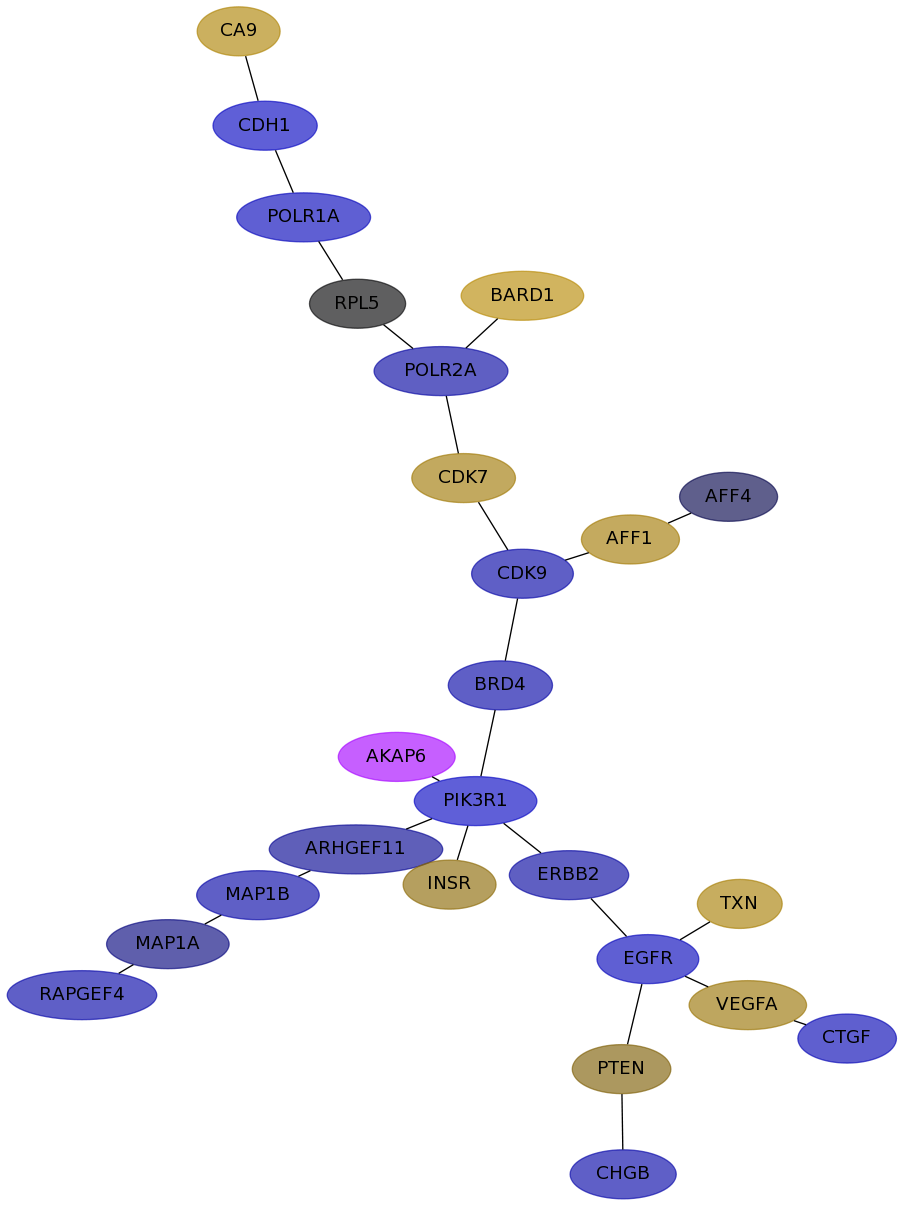
Score for each gene in subnetwork 6896 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| CHGB |   | 7 | 256 | 682 | 664 | -0.126 | -0.041 | 0.018 | 0.102 | -0.131 |
|---|
| BRD4 |   | 11 | 148 | 1047 | 1003 | -0.119 | 0.095 | -0.075 | 0.081 | -0.044 |
|---|
| TXN |   | 2 | 743 | 1486 | 1473 | 0.124 | 0.012 | 0.014 | 0.006 | -0.006 |
|---|
| ARHGEF11 |   | 1 | 1195 | 1598 | 1616 | -0.075 | -0.176 | -0.001 | 0.226 | -0.156 |
|---|
| POLR1A |   | 6 | 301 | 457 | 452 | -0.193 | -0.066 | undef | 0.093 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| RAPGEF4 |   | 1 | 1195 | 1598 | 1616 | -0.125 | 0.191 | -0.034 | 0.186 | 0.125 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDK7 |   | 7 | 256 | 318 | 328 | 0.105 | 0.184 | 0.017 | 0.078 | -0.097 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| RPL5 |   | 1 | 1195 | 1598 | 1616 | -0.000 | 0.128 | 0.220 | -0.194 | -0.110 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| POLR2A |   | 6 | 301 | 1547 | 1510 | -0.107 | 0.247 | 0.091 | 0.040 | 0.141 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| AKAP6 |   | 3 | 557 | 882 | 871 | undef | 0.116 | -0.125 | 0.026 | 0.153 |
|---|
| AFF4 |   | 1 | 1195 | 1598 | 1616 | -0.014 | 0.118 | -0.113 | 0.251 | undef |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| MAP1A |   | 12 | 140 | 1 | 36 | -0.045 | 0.143 | -0.054 | 0.162 | 0.147 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| CDK9 |   | 3 | 557 | 1598 | 1574 | -0.120 | 0.179 | 0.182 | 0.186 | -0.135 |
|---|
| AFF1 |   | 2 | 743 | 1598 | 1586 | 0.110 | 0.059 | -0.063 | 0.187 | 0.147 |
|---|
| CA9 |   | 2 | 743 | 882 | 892 | 0.146 | 0.137 | 0.068 | 0.099 | 0.116 |
|---|
GO Enrichment output for subnetwork 6896 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.249E-10 | 3.051E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.283E-10 | 4.011E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.74E-10 | 3.046E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 8.712E-10 | 5.321E-07 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.739E-09 | 8.499E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.836E-09 | 1.562E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 5.2E-09 | 1.815E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 8.157E-09 | 2.491E-06 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 1.761E-08 | 4.781E-06 |
|---|
| cardiac muscle tissue development | GO:0048738 |  | 5.822E-08 | 1.422E-05 |
|---|
| endothelial cell migration | GO:0043542 |  | 5.822E-08 | 1.293E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.676E-10 | 4.031E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 4.72E-10 | 5.678E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 5.017E-10 | 4.024E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.168E-09 | 7.029E-07 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.333E-09 | 1.122E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.879E-09 | 2.357E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 6.972E-09 | 2.396E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.094E-08 | 3.289E-06 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 2.361E-08 | 6.311E-06 |
|---|
| endothelial cell migration | GO:0043542 |  | 7.799E-08 | 1.877E-05 |
|---|
| inositol metabolic process | GO:0006020 |  | 7.799E-08 | 1.706E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.227E-08 | 2.823E-05 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 2.881E-08 | 3.313E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.193E-07 | 1.681E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.316E-07 | 1.906E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 4.376E-07 | 2.013E-04 |
|---|
| protein heterooligomerization | GO:0051291 |  | 7.643E-07 | 2.93E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 7.643E-07 | 2.511E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.22E-06 | 3.509E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.22E-06 | 3.119E-04 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.827E-06 | 4.202E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.605E-06 | 5.446E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.253E-06 | 2.309E-03 |
|---|
| regulation of cell migration | GO:0030334 |  | 1.817E-06 | 1.674E-03 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.035E-06 | 1.25E-03 |
|---|
| regulation of locomotion | GO:0040012 |  | 3.096E-06 | 1.426E-03 |
|---|
| regulation of cell motion | GO:0051270 |  | 3.096E-06 | 1.141E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.16E-06 | 9.705E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 3.547E-06 | 9.34E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 3.547E-06 | 8.173E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.982E-06 | 8.154E-04 |
|---|
| RNA elongation | GO:0006354 |  | 4.428E-06 | 8.161E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 4.924E-06 | 8.25E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.227E-08 | 2.823E-05 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 2.881E-08 | 3.313E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.193E-07 | 1.681E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.316E-07 | 1.906E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 4.376E-07 | 2.013E-04 |
|---|
| protein heterooligomerization | GO:0051291 |  | 7.643E-07 | 2.93E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 7.643E-07 | 2.511E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.22E-06 | 3.509E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.22E-06 | 3.119E-04 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.827E-06 | 4.202E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.605E-06 | 5.446E-04 |
|---|



