Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6893
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7489 | 3.838e-03 | 6.400e-04 | 7.827e-01 | 1.923e-06 |
|---|
| Loi | 0.2306 | 7.977e-02 | 1.123e-02 | 4.270e-01 | 3.826e-04 |
|---|
| Schmidt | 0.6762 | 0.000e+00 | 0.000e+00 | 4.199e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.431e-03 | 0.000e+00 |
|---|
| Wang | 0.2636 | 2.415e-03 | 4.136e-02 | 3.847e-01 | 3.843e-05 |
|---|
Expression data for subnetwork 6893 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
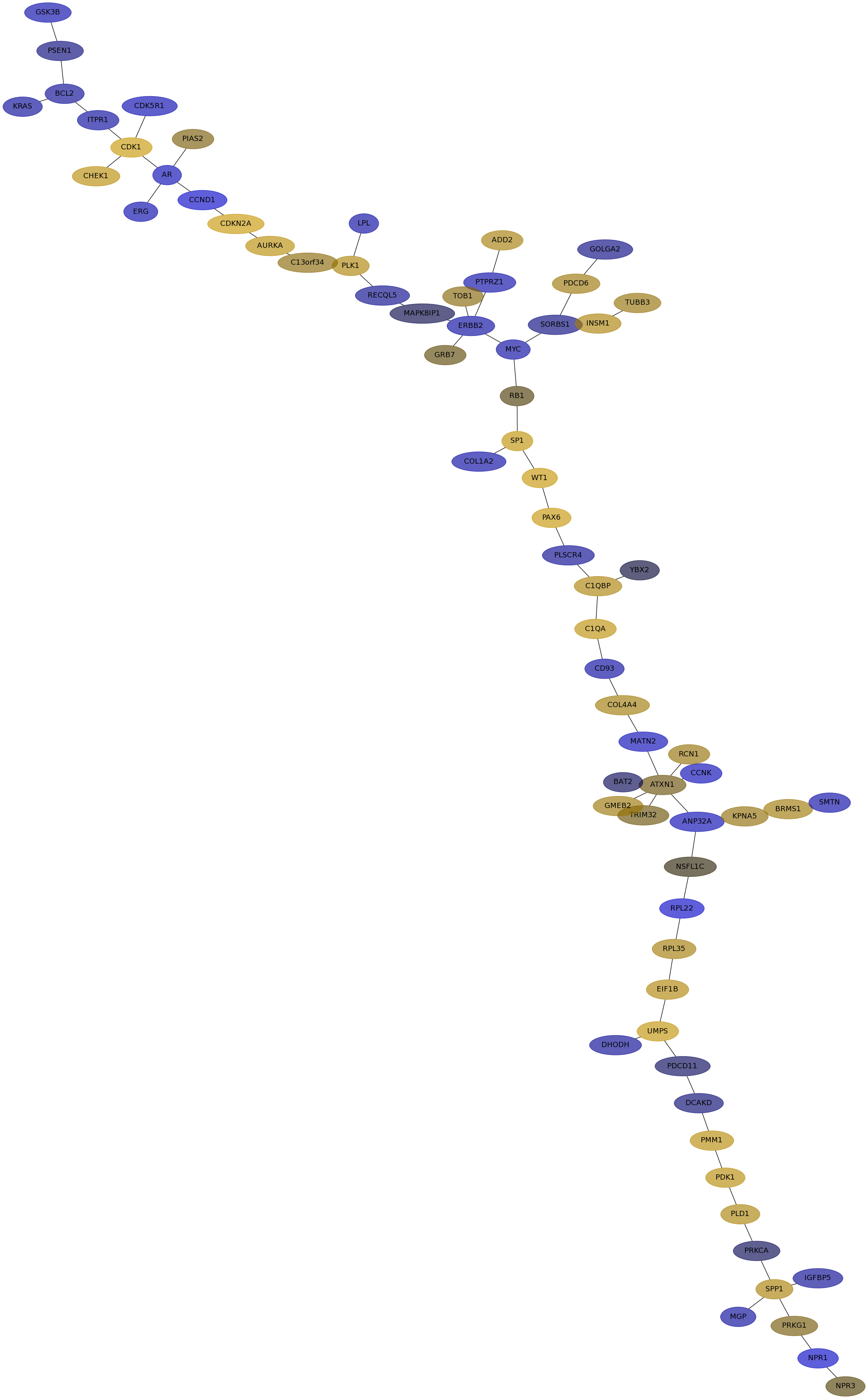
Score for each gene in subnetwork 6893 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| DCAKD |   | 1 | 1195 | 1576 | 1594 | -0.033 | -0.131 | -0.104 | undef | -0.250 |
|---|
| MGP |   | 3 | 557 | 1450 | 1430 | -0.088 | 0.007 | 0.150 | -0.084 | -0.030 |
|---|
| ADD2 |   | 8 | 222 | 614 | 599 | 0.113 | -0.022 | -0.061 | 0.270 | 0.180 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| EIF1B |   | 8 | 222 | 1576 | 1529 | 0.149 | 0.075 | 0.332 | -0.017 | -0.070 |
|---|
| RCN1 |   | 1 | 1195 | 1576 | 1594 | 0.076 | 0.109 | -0.012 | -0.010 | -0.042 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| NSFL1C |   | 2 | 743 | 1576 | 1561 | 0.005 | 0.016 | -0.204 | 0.137 | -0.021 |
|---|
| RECQL5 |   | 2 | 743 | 1165 | 1163 | -0.061 | -0.061 | -0.027 | 0.091 | -0.252 |
|---|
| PLK1 |   | 11 | 148 | 236 | 232 | 0.142 | -0.143 | 0.094 | -0.192 | -0.163 |
|---|
| RPL35 |   | 8 | 222 | 1323 | 1280 | 0.108 | -0.012 | 0.370 | -0.145 | -0.220 |
|---|
| KPNA5 |   | 1 | 1195 | 1576 | 1594 | 0.071 | -0.024 | 0.285 | -0.035 | 0.123 |
|---|
| PDK1 |   | 10 | 167 | 1 | 40 | 0.183 | -0.138 | -0.041 | -0.265 | -0.126 |
|---|
| YBX2 |   | 1 | 1195 | 1576 | 1594 | -0.007 | -0.046 | -0.079 | 0.125 | 0.082 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| BRMS1 |   | 1 | 1195 | 1576 | 1594 | 0.097 | -0.078 | -0.154 | 0.068 | -0.245 |
|---|
| PMM1 |   | 1 | 1195 | 1576 | 1594 | 0.177 | 0.063 | 0.125 | 0.005 | 0.048 |
|---|
| C1QA |   | 1 | 1195 | 1576 | 1594 | 0.205 | -0.229 | -0.116 | -0.159 | -0.037 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| TRIM32 |   | 1 | 1195 | 1576 | 1594 | 0.029 | 0.097 | 0.283 | -0.024 | 0.067 |
|---|
| RPL22 |   | 3 | 557 | 1576 | 1548 | -0.266 | 0.105 | 0.059 | -0.143 | 0.112 |
|---|
| PDCD11 |   | 2 | 743 | 1576 | 1561 | -0.019 | 0.075 | 0.091 | undef | 0.114 |
|---|
| CD93 |   | 2 | 743 | 842 | 854 | -0.102 | 0.036 | 0.160 | -0.146 | 0.254 |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| DHODH |   | 1 | 1195 | 1576 | 1594 | -0.074 | -0.186 | 0.067 | undef | -0.021 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| PSEN1 |   | 10 | 167 | 366 | 362 | -0.041 | 0.110 | undef | 0.083 | undef |
|---|
| C1QBP |   | 2 | 743 | 1010 | 1007 | 0.134 | -0.072 | 0.080 | -0.072 | -0.006 |
|---|
| CCNK |   | 1 | 1195 | 1576 | 1594 | -0.172 | -0.207 | -0.119 | undef | 0.084 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| NPR3 |   | 1 | 1195 | 1576 | 1594 | 0.016 | 0.005 | 0.195 | 0.034 | 0.034 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| PRKG1 |   | 3 | 557 | 179 | 213 | 0.035 | -0.125 | -0.074 | 0.183 | -0.095 |
|---|
| SMTN |   | 1 | 1195 | 1576 | 1594 | -0.115 | 0.319 | 0.042 | undef | 0.093 |
|---|
| PTPRZ1 |   | 9 | 196 | 570 | 554 | -0.114 | 0.183 | 0.233 | -0.316 | -0.095 |
|---|
| ANP32A |   | 1 | 1195 | 1576 | 1594 | -0.163 | -0.031 | 0.177 | undef | -0.006 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| GMEB2 |   | 1 | 1195 | 1576 | 1594 | 0.091 | 0.035 | 0.207 | -0.183 | 0.129 |
|---|
| TUBB3 |   | 7 | 256 | 764 | 756 | 0.078 | 0.130 | 0.073 | 0.207 | -0.079 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| ERG |   | 9 | 196 | 179 | 195 | -0.134 | -0.026 | 0.235 | -0.027 | 0.182 |
|---|
| BAT2 |   | 3 | 557 | 957 | 949 | -0.017 | 0.031 | 0.058 | undef | 0.016 |
|---|
| COL4A4 |   | 6 | 301 | 83 | 105 | 0.102 | 0.155 | -0.039 | 0.295 | 0.068 |
|---|
| NPR1 |   | 1 | 1195 | 1576 | 1594 | -0.259 | -0.122 | 0.191 | -0.041 | -0.180 |
|---|
| KRAS |   | 2 | 743 | 1510 | 1502 | -0.083 | -0.127 | -0.030 | undef | 0.182 |
|---|
| C13orf34 |   | 3 | 557 | 1165 | 1151 | 0.059 | 0.057 | 0.007 | undef | -0.147 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| MAPK8IP1 |   | 4 | 440 | 1576 | 1543 | -0.012 | 0.058 | -0.020 | 0.282 | -0.049 |
|---|
| PDCD6 |   | 10 | 167 | 1 | 40 | 0.091 | -0.060 | undef | undef | undef |
|---|
| GOLGA2 |   | 2 | 743 | 1492 | 1474 | -0.048 | 0.213 | 0.050 | undef | 0.052 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| ITPR1 |   | 4 | 440 | 1576 | 1543 | -0.096 | 0.077 | 0.015 | undef | 0.008 |
|---|
| MATN2 |   | 1 | 1195 | 1576 | 1594 | -0.164 | 0.026 | 0.160 | undef | -0.073 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| PIAS2 |   | 2 | 743 | 614 | 632 | 0.039 | -0.072 | 0.194 | 0.046 | -0.172 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| AURKA |   | 3 | 557 | 1165 | 1151 | 0.189 | -0.176 | 0.100 | -0.051 | 0.050 |
|---|
| UMPS |   | 3 | 557 | 842 | 834 | 0.229 | 0.047 | 0.080 | -0.098 | 0.021 |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| COL1A2 |   | 7 | 256 | 552 | 542 | -0.110 | 0.207 | 0.046 | 0.235 | 0.097 |
|---|
| TOB1 |   | 6 | 301 | 525 | 517 | 0.055 | 0.097 | -0.069 | 0.107 | -0.081 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6893 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.915E-08 | 7.122E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.261E-06 | 1.54E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.054E-06 | 3.301E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.526E-06 | 3.375E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 5.686E-06 | 2.778E-03 |
|---|
| response to metal ion | GO:0010038 |  | 7.404E-06 | 3.015E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 7.586E-06 | 2.647E-03 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 8.391E-06 | 2.562E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.739E-06 | 2.372E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 9.061E-06 | 2.214E-03 |
|---|
| response to inorganic substance | GO:0010035 |  | 1.103E-05 | 2.451E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.104E-08 | 9.874E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.425E-06 | 1.714E-03 |
|---|
| interphase | GO:0051325 |  | 1.758E-06 | 1.41E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.962E-06 | 2.985E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 6.635E-06 | 3.193E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.07E-06 | 2.835E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.057E-05 | 3.633E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.069E-05 | 3.214E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.49E-05 | 3.984E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.579E-05 | 3.799E-03 |
|---|
| pyrimidine base metabolic process | GO:0006206 |  | 1.579E-05 | 3.453E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.883E-06 | 4.331E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 7.749E-06 | 8.911E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 1.313E-05 | 0.01006982 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.313E-05 | 7.552E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.498E-05 | 6.892E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.09E-05 | 8.013E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 2.09E-05 | 6.868E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.18E-05 | 6.266E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.079E-05 | 7.868E-03 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 3.119E-05 | 7.173E-03 |
|---|
| pyrimidine base metabolic process | GO:0006206 |  | 3.119E-05 | 6.521E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| focal adhesion formation | GO:0048041 |  | 4.49E-06 | 8.275E-03 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 4.49E-06 | 4.138E-03 |
|---|
| peptidyl-serine modification | GO:0018209 |  | 1.246E-05 | 7.656E-03 |
|---|
| epidermis morphogenesis | GO:0048730 |  | 2.647E-05 | 0.012198 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 2.647E-05 | 9.758E-03 |
|---|
| posttranscriptional regulation of gene expression | GO:0010608 |  | 2.852E-05 | 8.759E-03 |
|---|
| regulation of ossification | GO:0030278 |  | 3.222E-05 | 8.483E-03 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 4.189E-05 | 9.651E-03 |
|---|
| negative regulation of cell size | GO:0045792 |  | 4.746E-05 | 9.718E-03 |
|---|
| renal system process | GO:0003014 |  | 4.811E-05 | 8.867E-03 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 5.354E-05 | 8.971E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.883E-06 | 4.331E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 7.749E-06 | 8.911E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 1.313E-05 | 0.01006982 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.313E-05 | 7.552E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.498E-05 | 6.892E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.09E-05 | 8.013E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 2.09E-05 | 6.868E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.18E-05 | 6.266E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.079E-05 | 7.868E-03 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 3.119E-05 | 7.173E-03 |
|---|
| pyrimidine base metabolic process | GO:0006206 |  | 3.119E-05 | 6.521E-03 |
|---|



