Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6880
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7489 | 3.838e-03 | 6.400e-04 | 7.828e-01 | 1.923e-06 |
|---|
| Loi | 0.2310 | 7.917e-02 | 1.107e-02 | 4.255e-01 | 3.730e-04 |
|---|
| Schmidt | 0.6760 | 0.000e+00 | 0.000e+00 | 4.223e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.427e-03 | 0.000e+00 |
|---|
| Wang | 0.2637 | 2.410e-03 | 4.131e-02 | 3.844e-01 | 3.827e-05 |
|---|
Expression data for subnetwork 6880 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
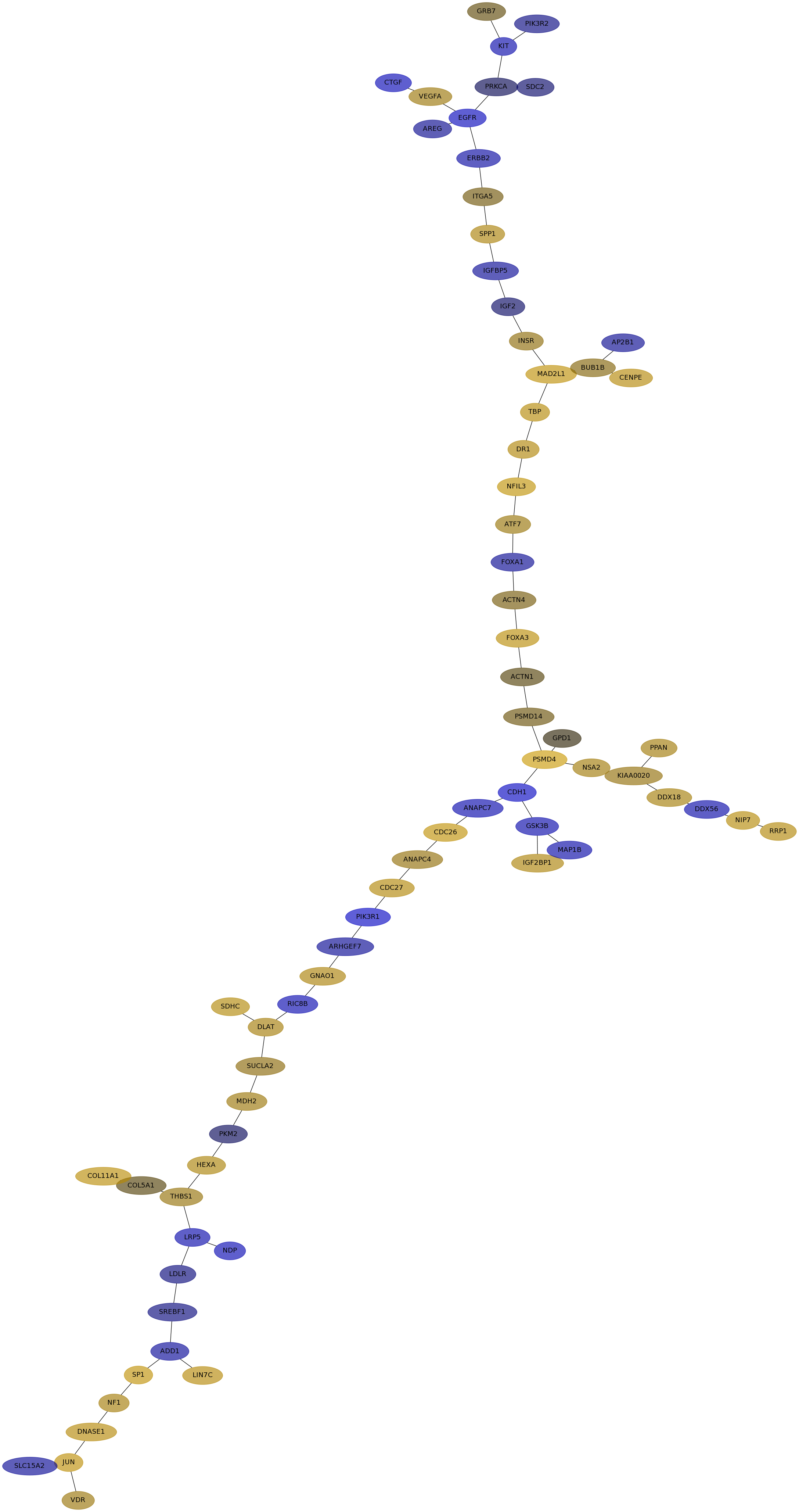
Score for each gene in subnetwork 6880 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| ADD1 |   | 8 | 222 | 366 | 373 | -0.085 | 0.116 | 0.035 | undef | -0.098 |
|---|
| DDX18 |   | 1 | 1195 | 1141 | 1170 | 0.112 | -0.142 | 0.000 | 0.043 | 0.178 |
|---|
| NF1 |   | 9 | 196 | 552 | 541 | 0.110 | -0.052 | -0.032 | 0.090 | -0.094 |
|---|
| MDH2 |   | 1 | 1195 | 1141 | 1170 | 0.089 | 0.180 | 0.044 | 0.120 | -0.008 |
|---|
| LDLR |   | 5 | 360 | 780 | 767 | -0.041 | 0.218 | 0.039 | -0.028 | 0.064 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| COL11A1 |   | 3 | 557 | 525 | 540 | 0.191 | 0.149 | -0.023 | undef | 0.309 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| PIK3R2 |   | 4 | 440 | 141 | 165 | -0.047 | 0.015 | -0.147 | undef | -0.135 |
|---|
| COL5A1 |   | 2 | 743 | 1141 | 1144 | 0.016 | 0.226 | 0.006 | 0.075 | 0.201 |
|---|
| PSMD14 |   | 4 | 440 | 842 | 831 | 0.027 | 0.135 | -0.179 | -0.018 | -0.031 |
|---|
| AP2B1 |   | 6 | 301 | 780 | 764 | -0.070 | -0.106 | 0.070 | 0.252 | 0.094 |
|---|
| ANAPC7 |   | 1 | 1195 | 1141 | 1170 | -0.135 | -0.242 | undef | 0.092 | undef |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| DNASE1 |   | 4 | 440 | 590 | 585 | 0.165 | -0.301 | -0.088 | -0.078 | 0.114 |
|---|
| RRP1 |   | 1 | 1195 | 1141 | 1170 | 0.154 | -0.104 | 0.095 | undef | -0.135 |
|---|
| BUB1B |   | 4 | 440 | 1010 | 991 | 0.050 | -0.164 | 0.037 | -0.101 | 0.148 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| LRP5 |   | 3 | 557 | 1141 | 1129 | -0.129 | -0.051 | 0.011 | 0.017 | -0.146 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| KIAA0020 |   | 2 | 743 | 1141 | 1144 | 0.071 | -0.112 | 0.018 | -0.140 | -0.017 |
|---|
| NSA2 |   | 4 | 440 | 806 | 790 | 0.103 | 0.097 | 0.132 | -0.026 | 0.006 |
|---|
| VDR |   | 3 | 557 | 1141 | 1129 | 0.086 | 0.208 | -0.003 | -0.051 | -0.046 |
|---|
| AREG |   | 5 | 360 | 1141 | 1109 | -0.067 | 0.112 | 0.076 | -0.173 | -0.054 |
|---|
| NIP7 |   | 1 | 1195 | 1141 | 1170 | 0.168 | -0.241 | 0.008 | -0.111 | 0.059 |
|---|
| DR1 |   | 3 | 557 | 1141 | 1129 | 0.151 | 0.041 | 0.174 | -0.054 | 0.104 |
|---|
| MAD2L1 |   | 8 | 222 | 236 | 236 | 0.208 | -0.130 | 0.116 | 0.032 | 0.063 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| DLAT |   | 4 | 440 | 1141 | 1127 | 0.111 | 0.029 | 0.072 | undef | -0.014 |
|---|
| SDHC |   | 3 | 557 | 1141 | 1129 | 0.158 | -0.033 | -0.077 | 0.199 | 0.110 |
|---|
| TBP |   | 4 | 440 | 1141 | 1127 | 0.171 | 0.017 | 0.009 | -0.061 | -0.009 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| RIC8B |   | 1 | 1195 | 1141 | 1170 | -0.148 | -0.091 | 0.119 | 0.009 | -0.001 |
|---|
| FOXA3 |   | 12 | 140 | 318 | 322 | 0.192 | 0.289 | undef | -0.171 | undef |
|---|
| SDC2 |   | 4 | 440 | 590 | 585 | -0.027 | 0.068 | 0.033 | undef | 0.215 |
|---|
| GPD1 |   | 7 | 256 | 658 | 644 | 0.006 | -0.019 | 0.112 | -0.034 | 0.033 |
|---|
| THBS1 |   | 5 | 360 | 1141 | 1109 | 0.078 | 0.044 | 0.185 | 0.200 | 0.267 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| DDX56 |   | 1 | 1195 | 1141 | 1170 | -0.119 | 0.102 | -0.220 | 0.085 | -0.107 |
|---|
| SREBF1 |   | 9 | 196 | 478 | 460 | -0.042 | 0.232 | -0.195 | 0.140 | -0.074 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PPAN |   | 1 | 1195 | 1141 | 1170 | 0.104 | -0.002 | -0.156 | -0.185 | -0.118 |
|---|
| SUCLA2 |   | 1 | 1195 | 1141 | 1170 | 0.058 | -0.087 | 0.000 | undef | 0.126 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| PKM2 |   | 1 | 1195 | 1141 | 1170 | -0.018 | 0.257 | 0.056 | undef | -0.050 |
|---|
| PSMD4 |   | 11 | 148 | 296 | 282 | 0.285 | -0.243 | -0.043 | 0.087 | -0.045 |
|---|
| CDC26 |   | 2 | 743 | 1083 | 1084 | 0.219 | -0.108 | undef | undef | undef |
|---|
| LIN7C |   | 10 | 167 | 590 | 566 | 0.166 | 0.088 | 0.118 | -0.018 | 0.094 |
|---|
| SLC15A2 |   | 4 | 440 | 928 | 912 | -0.076 | 0.109 | -0.068 | 0.050 | -0.123 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CDC27 |   | 6 | 301 | 1010 | 986 | 0.156 | -0.086 | -0.029 | 0.006 | 0.348 |
|---|
| NDP |   | 1 | 1195 | 1141 | 1170 | -0.158 | -0.044 | -0.125 | -0.109 | 0.012 |
|---|
| GNAO1 |   | 3 | 557 | 478 | 487 | 0.129 | 0.025 | -0.095 | undef | 0.173 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| NFIL3 |   | 3 | 557 | 1141 | 1129 | 0.222 | -0.163 | -0.095 | -0.029 | 0.130 |
|---|
| CENPE |   | 1 | 1195 | 1141 | 1170 | 0.165 | -0.102 | 0.150 | 0.032 | 0.040 |
|---|
| HEXA |   | 3 | 557 | 1141 | 1129 | 0.131 | -0.121 | 0.091 | -0.031 | -0.144 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
| IGF2 |   | 14 | 117 | 570 | 553 | -0.022 | 0.172 | 0.145 | 0.087 | 0.116 |
|---|
| ANAPC4 |   | 3 | 557 | 1083 | 1073 | 0.071 | 0.091 | undef | 0.030 | undef |
|---|
GO Enrichment output for subnetwork 6880 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.647E-13 | 4.023E-10 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.955E-13 | 2.388E-10 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.955E-13 | 1.592E-10 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 3.777E-13 | 2.307E-10 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.091E-12 | 5.329E-10 |
|---|
| regulation of ligase activity | GO:0051340 |  | 2.186E-12 | 8.903E-10 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 6.861E-12 | 2.395E-09 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.382E-10 | 7.274E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 3.525E-10 | 9.568E-08 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 6.518E-10 | 1.592E-07 |
|---|
| acetyl-CoA metabolic process | GO:0006084 |  | 8.888E-09 | 1.974E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 3.521E-13 | 8.471E-10 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.178E-13 | 5.026E-10 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.178E-13 | 3.35E-10 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.062E-13 | 4.849E-10 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.322E-12 | 1.117E-09 |
|---|
| regulation of ligase activity | GO:0051340 |  | 4.647E-12 | 1.863E-09 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.454E-11 | 4.996E-09 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 4.433E-10 | 1.333E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 6.553E-10 | 1.752E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.21E-09 | 2.91E-07 |
|---|
| acetyl-CoA metabolic process | GO:0006084 |  | 1.063E-08 | 2.326E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.356E-08 | 5.418E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.693E-08 | 3.097E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.693E-08 | 2.065E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.495E-08 | 2.584E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 9.102E-08 | 4.187E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.405E-07 | 5.387E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 2.336E-07 | 7.674E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.614E-07 | 7.515E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.614E-07 | 6.68E-05 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 4.164E-07 | 9.577E-05 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 5.43E-07 | 1.135E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell motion | GO:0051270 |  | 7.666E-08 | 1.413E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.942E-07 | 2.711E-04 |
|---|
| regulation of blood vessel endothelial cell migration | GO:0043535 |  | 3.029E-07 | 1.861E-04 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 3.897E-07 | 1.796E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.652E-07 | 1.715E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.652E-07 | 1.429E-04 |
|---|
| regulation of cell migration | GO:0030334 |  | 6.474E-07 | 1.705E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.126E-07 | 1.872E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.053E-06 | 2.156E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.193E-06 | 2.199E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.193E-06 | 1.999E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.356E-08 | 5.418E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.693E-08 | 3.097E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.693E-08 | 2.065E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.495E-08 | 2.584E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 9.102E-08 | 4.187E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.405E-07 | 5.387E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 2.336E-07 | 7.674E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.614E-07 | 7.515E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.614E-07 | 6.68E-05 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 4.164E-07 | 9.577E-05 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 5.43E-07 | 1.135E-04 |
|---|



