Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6877
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7489 | 3.840e-03 | 6.410e-04 | 7.828e-01 | 1.927e-06 |
|---|
| Loi | 0.2309 | 7.938e-02 | 1.113e-02 | 4.261e-01 | 3.764e-04 |
|---|
| Schmidt | 0.6761 | 0.000e+00 | 0.000e+00 | 4.212e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.420e-03 | 0.000e+00 |
|---|
| Wang | 0.2636 | 2.413e-03 | 4.134e-02 | 3.846e-01 | 3.837e-05 |
|---|
Expression data for subnetwork 6877 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
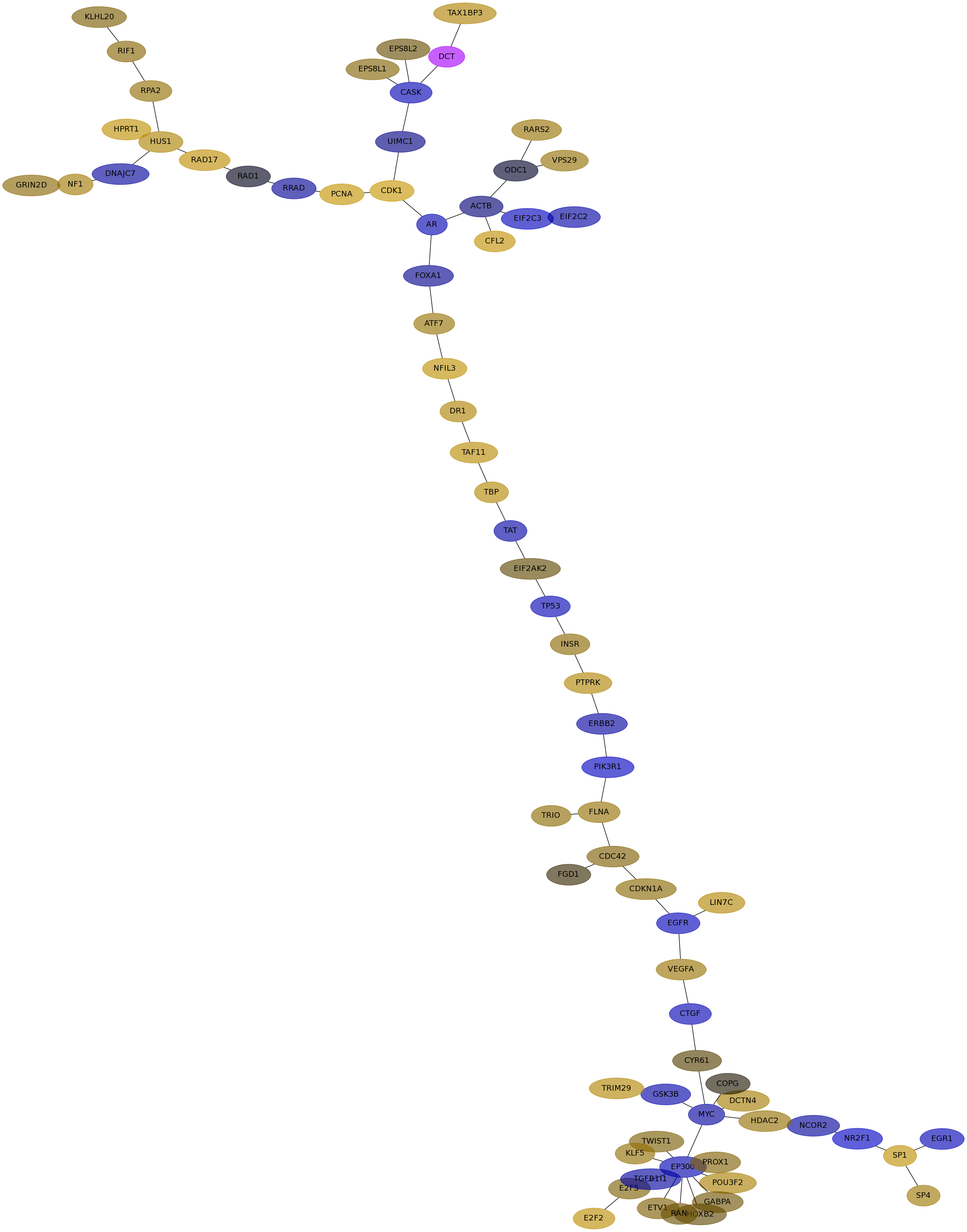
Score for each gene in subnetwork 6877 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| NF1 |   | 9 | 196 | 552 | 541 | 0.110 | -0.052 | -0.032 | 0.090 | -0.094 |
|---|
| EPS8L1 |   | 2 | 743 | 1274 | 1260 | 0.054 | 0.202 | -0.095 | undef | 0.029 |
|---|
| RARS2 |   | 1 | 1195 | 1274 | 1296 | 0.086 | -0.031 | undef | undef | undef |
|---|
| E2F5 |   | 2 | 743 | 1274 | 1260 | 0.048 | -0.087 | 0.002 | -0.032 | -0.074 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| RRAD |   | 2 | 743 | 1034 | 1039 | -0.081 | 0.171 | -0.082 | 0.069 | 0.117 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| EPS8L2 |   | 2 | 743 | 478 | 490 | 0.028 | 0.287 | -0.160 | undef | 0.106 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| EIF2AK2 |   | 9 | 196 | 648 | 629 | 0.023 | -0.022 | -0.134 | 0.096 | -0.303 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| TAX1BP3 |   | 2 | 743 | 1274 | 1260 | 0.150 | 0.219 | 0.070 | undef | 0.206 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| RAN |   | 3 | 557 | 236 | 265 | 0.030 | -0.006 | 0.062 | 0.016 | undef |
|---|
| TAF11 |   | 3 | 557 | 457 | 463 | 0.192 | 0.112 | 0.237 | 0.053 | 0.180 |
|---|
| ETV1 |   | 10 | 167 | 1 | 40 | 0.046 | 0.054 | -0.065 | 0.014 | 0.141 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
| TAT |   | 11 | 148 | 83 | 97 | -0.113 | 0.030 | -0.182 | 0.037 | -0.074 |
|---|
| RAD1 |   | 2 | 743 | 1274 | 1260 | -0.003 | -0.168 | 0.120 | undef | 0.001 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| PROX1 |   | 4 | 440 | 614 | 618 | 0.050 | -0.030 | -0.128 | 0.161 | 0.112 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| RAD17 |   | 1 | 1195 | 1274 | 1296 | 0.214 | 0.064 | 0.051 | undef | 0.061 |
|---|
| DCT |   | 1 | 1195 | 1274 | 1296 | undef | -0.019 | 0.000 | -0.087 | -0.092 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| ODC1 |   | 4 | 440 | 141 | 165 | -0.006 | -0.099 | 0.209 | -0.207 | -0.107 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| SP4 |   | 1 | 1195 | 1274 | 1296 | 0.102 | -0.235 | -0.088 | -0.134 | 0.199 |
|---|
| HUS1 |   | 1 | 1195 | 1274 | 1296 | 0.148 | 0.074 | -0.158 | undef | 0.153 |
|---|
| HPRT1 |   | 1 | 1195 | 1274 | 1296 | 0.216 | -0.215 | 0.100 | -0.034 | 0.020 |
|---|
| DR1 |   | 3 | 557 | 1141 | 1129 | 0.151 | 0.041 | 0.174 | -0.054 | 0.104 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| UIMC1 |   | 6 | 301 | 179 | 205 | -0.054 | 0.226 | 0.031 | -0.005 | 0.189 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| ACTB |   | 3 | 557 | 1274 | 1258 | -0.039 | 0.283 | -0.061 | 0.164 | 0.003 |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| HDAC2 |   | 13 | 124 | 318 | 316 | 0.079 | -0.094 | 0.148 | 0.089 | 0.037 |
|---|
| COPG |   | 5 | 360 | 570 | 562 | 0.004 | 0.080 | -0.158 | undef | 0.280 |
|---|
| FGD1 |   | 2 | 743 | 1274 | 1260 | 0.008 | -0.131 | -0.053 | 0.043 | -0.074 |
|---|
| TRIM29 |   | 8 | 222 | 296 | 287 | 0.161 | 0.240 | -0.150 | -0.077 | 0.024 |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| TBP |   | 4 | 440 | 1141 | 1127 | 0.171 | 0.017 | 0.009 | -0.061 | -0.009 |
|---|
| EIF2C2 |   | 1 | 1195 | 1274 | 1296 | -0.117 | -0.099 | 0.159 | undef | 0.113 |
|---|
| RIF1 |   | 2 | 743 | 552 | 568 | 0.057 | -0.060 | -0.035 | undef | -0.092 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| TRIO |   | 3 | 557 | 366 | 389 | 0.067 | 0.058 | -0.062 | 0.196 | -0.134 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| RPA2 |   | 1 | 1195 | 1274 | 1296 | 0.075 | -0.056 | 0.082 | 0.117 | 0.067 |
|---|
| KLHL20 |   | 2 | 743 | 1274 | 1260 | 0.045 | 0.201 | -0.058 | 0.182 | 0.240 |
|---|
| CYR61 |   | 4 | 440 | 991 | 978 | 0.018 | 0.206 | 0.203 | 0.092 | 0.234 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| DCTN4 |   | 5 | 360 | 1010 | 987 | 0.122 | -0.025 | -0.111 | 0.173 | -0.105 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| DNAJC7 |   | 1 | 1195 | 1274 | 1296 | -0.095 | -0.120 | -0.069 | -0.214 | 0.174 |
|---|
| CASK |   | 2 | 743 | 1274 | 1260 | -0.170 | 0.160 | -0.077 | 0.139 | -0.166 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| LIN7C |   | 10 | 167 | 590 | 566 | 0.166 | 0.088 | 0.118 | -0.018 | 0.094 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| CFL2 |   | 1 | 1195 | 1274 | 1296 | 0.235 | 0.026 | undef | undef | undef |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| NFIL3 |   | 3 | 557 | 1141 | 1129 | 0.222 | -0.163 | -0.095 | -0.029 | 0.130 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GRIN2D |   | 13 | 124 | 552 | 522 | 0.058 | 0.155 | 0.000 | undef | undef |
|---|
| VPS29 |   | 1 | 1195 | 1274 | 1296 | 0.079 | -0.217 | undef | undef | undef |
|---|
| EIF2C3 |   | 1 | 1195 | 1274 | 1296 | -0.198 | 0.070 | 0.040 | undef | -0.125 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6877 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA damage checkpoint | GO:0000077 |  | 9.133E-10 | 2.231E-06 |
|---|
| regulation of DNA replication | GO:0006275 |  | 9.459E-10 | 1.155E-06 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 1.69E-09 | 1.376E-06 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 4.211E-09 | 2.572E-06 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 4.628E-09 | 2.261E-06 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 4.049E-08 | 1.649E-05 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 5.879E-08 | 2.052E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.557E-08 | 2.002E-05 |
|---|
| negative regulation of DNA replication initiation | GO:0032297 |  | 6.557E-08 | 1.78E-05 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 7.988E-08 | 1.951E-05 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.524E-07 | 3.384E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA damage checkpoint | GO:0000077 |  | 1.276E-09 | 3.07E-06 |
|---|
| regulation of DNA replication | GO:0006275 |  | 2.276E-09 | 2.738E-06 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 2.454E-09 | 1.968E-06 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 9.046E-09 | 5.441E-06 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.108E-08 | 5.331E-06 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 7.87E-08 | 3.156E-05 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 9.999E-08 | 3.437E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.031E-07 | 3.101E-05 |
|---|
| negative regulation of DNA replication initiation | GO:0032297 |  | 1.031E-07 | 2.757E-05 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.318E-07 | 3.171E-05 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 2.664E-07 | 5.826E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of DNA replication | GO:0006275 |  | 2.986E-09 | 6.867E-06 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.155E-09 | 3.628E-06 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 6.331E-09 | 4.854E-06 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 1.719E-08 | 9.882E-06 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.871E-08 | 8.607E-06 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 1.457E-07 | 5.586E-05 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 2.136E-07 | 7.017E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.324E-07 | 6.683E-05 |
|---|
| negative regulation of DNA replication initiation | GO:0032297 |  | 2.324E-07 | 5.94E-05 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.37E-07 | 5.451E-05 |
|---|
| regulation of DNA replication initiation | GO:0030174 |  | 4.159E-07 | 8.696E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.045E-06 | 1.926E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.307E-06 | 1.204E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 1.307E-06 | 8.029E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.47E-06 | 1.138E-03 |
|---|
| prostate gland development | GO:0030850 |  | 2.96E-06 | 1.091E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 2.96E-06 | 9.093E-04 |
|---|
| negative regulation of neuroblast proliferation | GO:0007406 |  | 2.96E-06 | 7.794E-04 |
|---|
| urogenital system development | GO:0001655 |  | 5.072E-06 | 1.169E-03 |
|---|
| ER overload response | GO:0006983 |  | 5.892E-06 | 1.207E-03 |
|---|
| regulation of bone resorption | GO:0045124 |  | 5.892E-06 | 1.086E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 5.892E-06 | 9.872E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of DNA replication | GO:0006275 |  | 2.986E-09 | 6.867E-06 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.155E-09 | 3.628E-06 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 6.331E-09 | 4.854E-06 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 1.719E-08 | 9.882E-06 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.871E-08 | 8.607E-06 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 1.457E-07 | 5.586E-05 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 2.136E-07 | 7.017E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.324E-07 | 6.683E-05 |
|---|
| negative regulation of DNA replication initiation | GO:0032297 |  | 2.324E-07 | 5.94E-05 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.37E-07 | 5.451E-05 |
|---|
| regulation of DNA replication initiation | GO:0030174 |  | 4.159E-07 | 8.696E-05 |
|---|



