Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6853
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7490 | 3.830e-03 | 6.390e-04 | 7.825e-01 | 1.915e-06 |
|---|
| Loi | 0.2308 | 7.943e-02 | 1.114e-02 | 4.262e-01 | 3.772e-04 |
|---|
| Schmidt | 0.6758 | 0.000e+00 | 0.000e+00 | 4.242e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.422e-03 | 0.000e+00 |
|---|
| Wang | 0.2635 | 2.427e-03 | 4.148e-02 | 3.852e-01 | 3.878e-05 |
|---|
Expression data for subnetwork 6853 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
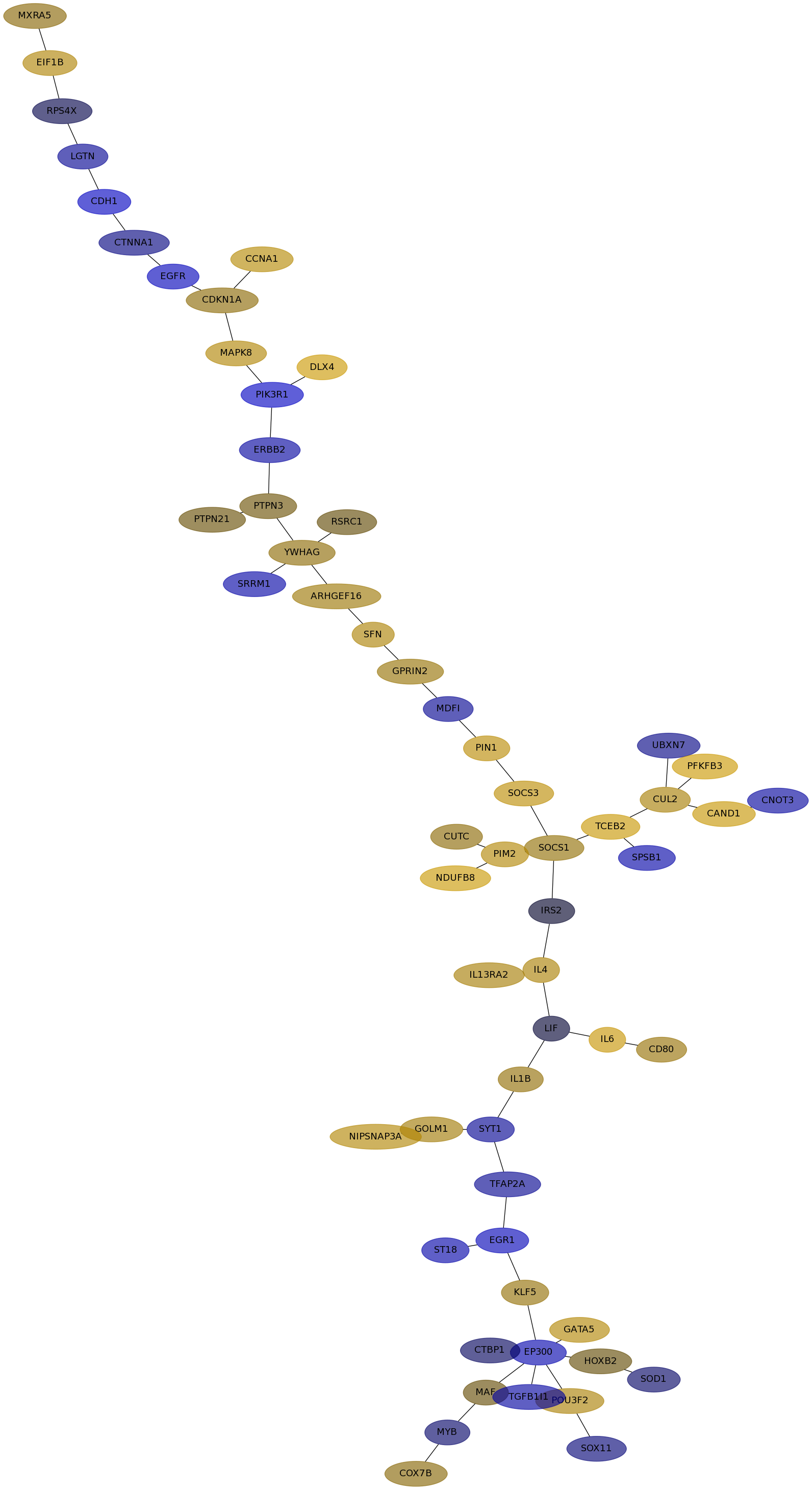
Score for each gene in subnetwork 6853 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| MAPK8 |   | 8 | 222 | 696 | 677 | 0.153 | 0.200 | 0.249 | -0.066 | 0.138 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| CTNNA1 |   | 2 | 743 | 1116 | 1121 | -0.049 | 0.218 | 0.010 | 0.115 | -0.098 |
|---|
| SRRM1 |   | 10 | 167 | 296 | 283 | -0.120 | 0.139 | 0.244 | 0.294 | -0.109 |
|---|
| PTPN21 |   | 5 | 360 | 1717 | 1677 | 0.027 | 0.177 | 0.214 | 0.205 | 0.119 |
|---|
| IL4 |   | 1 | 1195 | 1717 | 1730 | 0.135 | -0.060 | 0.000 | 0.038 | undef |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PTPN3 |   | 11 | 148 | 658 | 635 | 0.029 | 0.060 | -0.067 | 0.225 | -0.034 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| MXRA5 |   | 1 | 1195 | 1717 | 1730 | 0.058 | 0.220 | 0.018 | undef | 0.090 |
|---|
| SPSB1 |   | 3 | 557 | 1083 | 1073 | -0.124 | 0.088 | -0.152 | undef | 0.169 |
|---|
| EIF1B |   | 8 | 222 | 1576 | 1529 | 0.149 | 0.075 | 0.332 | -0.017 | -0.070 |
|---|
| IL1B |   | 1 | 1195 | 1717 | 1730 | 0.073 | -0.094 | 0.045 | -0.184 | -0.018 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| CUTC |   | 1 | 1195 | 1717 | 1730 | 0.063 | -0.086 | 0.150 | undef | 0.177 |
|---|
| CNOT3 |   | 1 | 1195 | 1717 | 1730 | -0.102 | 0.060 | -0.146 | 0.069 | -0.186 |
|---|
| ARHGEF16 |   | 10 | 167 | 296 | 283 | 0.096 | 0.036 | -0.047 | 0.200 | 0.038 |
|---|
| GOLM1 |   | 8 | 222 | 1534 | 1489 | 0.109 | 0.161 | -0.217 | 0.170 | -0.060 |
|---|
| UBXN7 |   | 3 | 557 | 727 | 728 | -0.055 | 0.153 | 0.224 | 0.167 | 0.175 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| SOCS1 |   | 3 | 557 | 1717 | 1689 | 0.074 | -0.153 | -0.177 | -0.202 | -0.100 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| TFAP2A |   | 3 | 557 | 83 | 117 | -0.070 | 0.146 | -0.095 | -0.152 | -0.009 |
|---|
| NDUFB8 |   | 5 | 360 | 696 | 680 | 0.286 | -0.012 | 0.340 | -0.171 | 0.205 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| COX7B |   | 1 | 1195 | 1717 | 1730 | 0.057 | -0.072 | 0.112 | -0.100 | 0.046 |
|---|
| DLX4 |   | 10 | 167 | 236 | 234 | 0.291 | 0.057 | -0.004 | undef | -0.059 |
|---|
| MAF |   | 1 | 1195 | 1717 | 1730 | 0.024 | 0.085 | -0.094 | -0.015 | 0.122 |
|---|
| SOX11 |   | 5 | 360 | 1396 | 1378 | -0.039 | -0.073 | -0.028 | 0.274 | -0.104 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| LIF |   | 1 | 1195 | 1717 | 1730 | -0.007 | 0.040 | 0.207 | -0.059 | -0.070 |
|---|
| SOD1 |   | 2 | 743 | 1717 | 1701 | -0.026 | -0.147 | 0.026 | 0.135 | -0.113 |
|---|
| TCEB2 |   | 2 | 743 | 1083 | 1084 | 0.281 | 0.005 | -0.072 | -0.025 | -0.002 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| PFKFB3 |   | 3 | 557 | 1659 | 1632 | 0.301 | 0.085 | 0.328 | 0.056 | 0.331 |
|---|
| CD80 |   | 4 | 440 | 1055 | 1035 | 0.081 | -0.199 | -0.136 | 0.034 | -0.078 |
|---|
| SYT1 |   | 11 | 148 | 1 | 39 | -0.079 | 0.051 | 0.038 | 0.189 | -0.124 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| CAND1 |   | 1 | 1195 | 1717 | 1730 | 0.266 | 0.059 | 0.109 | 0.077 | 0.106 |
|---|
| SOCS3 |   | 2 | 743 | 1717 | 1701 | 0.201 | -0.110 | 0.064 | 0.023 | -0.138 |
|---|
| IRS2 |   | 3 | 557 | 1102 | 1098 | -0.006 | 0.043 | 0.054 | -0.256 | 0.125 |
|---|
| RPS4X |   | 1 | 1195 | 1717 | 1730 | -0.014 | 0.081 | 0.183 | -0.098 | -0.068 |
|---|
| IL6 |   | 9 | 196 | 366 | 365 | 0.257 | -0.106 | 0.226 | 0.024 | 0.035 |
|---|
| NIPSNAP3A |   | 1 | 1195 | 1717 | 1730 | 0.160 | -0.121 | undef | undef | undef |
|---|
| CUL2 |   | 7 | 256 | 842 | 816 | 0.124 | -0.192 | -0.090 | -0.035 | 0.166 |
|---|
| LGTN |   | 1 | 1195 | 1717 | 1730 | -0.077 | 0.011 | 0.093 | undef | 0.125 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| PIM2 |   | 1 | 1195 | 1717 | 1730 | 0.162 | -0.275 | -0.152 | -0.253 | -0.102 |
|---|
| GATA5 |   | 8 | 222 | 366 | 373 | 0.161 | -0.031 | undef | undef | undef |
|---|
| CTBP1 |   | 3 | 557 | 991 | 982 | -0.022 | -0.021 | -0.298 | 0.196 | -0.143 |
|---|
| IL13RA2 |   | 1 | 1195 | 1717 | 1730 | 0.121 | 0.113 | -0.059 | 0.072 | 0.198 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
GO Enrichment output for subnetwork 6853 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 8.228E-09 | 2.01E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.169E-08 | 1.428E-05 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 3.621E-08 | 2.949E-05 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 4.255E-08 | 2.599E-05 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 4.295E-08 | 2.099E-05 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 4.891E-08 | 1.992E-05 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 6.895E-08 | 2.406E-05 |
|---|
| regulation of T cell activation | GO:0050863 |  | 1.181E-07 | 3.608E-05 |
|---|
| positive regulation of leukocyte proliferation | GO:0070665 |  | 4.725E-07 | 1.283E-04 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 5.196E-07 | 1.269E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 5.425E-07 | 1.205E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 9.489E-09 | 2.283E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.198E-08 | 1.441E-05 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 3.833E-08 | 3.074E-05 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 4.566E-08 | 2.747E-05 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 4.593E-08 | 2.21E-05 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 5.639E-08 | 2.261E-05 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 6.673E-08 | 2.294E-05 |
|---|
| regulation of T cell activation | GO:0050863 |  | 1.286E-07 | 3.868E-05 |
|---|
| positive regulation of leukocyte proliferation | GO:0070665 |  | 4.911E-07 | 1.313E-04 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 5.395E-07 | 1.298E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 6.25E-07 | 1.367E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 3.208E-08 | 7.378E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 5.994E-08 | 6.893E-05 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 1.145E-07 | 8.778E-05 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 1.258E-07 | 7.233E-05 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 1.532E-07 | 7.046E-05 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 1.58E-07 | 6.056E-05 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 2.371E-07 | 7.79E-05 |
|---|
| regulation of T cell activation | GO:0050863 |  | 6.63E-07 | 1.906E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 1.209E-06 | 3.089E-04 |
|---|
| positive regulation of leukocyte proliferation | GO:0070665 |  | 1.419E-06 | 3.264E-04 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.533E-06 | 3.206E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein amino acid phosphorylation | GO:0001932 |  | 1.465E-09 | 2.701E-06 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 4.878E-09 | 4.495E-06 |
|---|
| regulation of protein modification process | GO:0031399 |  | 1.087E-08 | 6.677E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 3.042E-07 | 1.401E-04 |
|---|
| regulation of tyrosine phosphorylation of Stat3 protein | GO:0042516 |  | 3.14E-07 | 1.157E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 6.746E-07 | 2.072E-04 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 9.21E-07 | 2.425E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 9.405E-07 | 2.167E-04 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 1.466E-06 | 3.002E-04 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 1.695E-06 | 3.124E-04 |
|---|
| regulation of tyrosine phosphorylation of STAT protein | GO:0042509 |  | 1.696E-06 | 2.841E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 3.208E-08 | 7.378E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 5.994E-08 | 6.893E-05 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 1.145E-07 | 8.778E-05 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 1.258E-07 | 7.233E-05 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 1.532E-07 | 7.046E-05 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 1.58E-07 | 6.056E-05 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 2.371E-07 | 7.79E-05 |
|---|
| regulation of T cell activation | GO:0050863 |  | 6.63E-07 | 1.906E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 1.209E-06 | 3.089E-04 |
|---|
| positive regulation of leukocyte proliferation | GO:0070665 |  | 1.419E-06 | 3.264E-04 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.533E-06 | 3.206E-04 |
|---|



