Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6832
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7495 | 3.799e-03 | 6.320e-04 | 7.814e-01 | 1.876e-06 |
|---|
| Loi | 0.2308 | 7.944e-02 | 1.114e-02 | 4.262e-01 | 3.773e-04 |
|---|
| Schmidt | 0.6757 | 0.000e+00 | 0.000e+00 | 4.254e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.431e-03 | 0.000e+00 |
|---|
| Wang | 0.2635 | 2.425e-03 | 4.146e-02 | 3.851e-01 | 3.872e-05 |
|---|
Expression data for subnetwork 6832 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
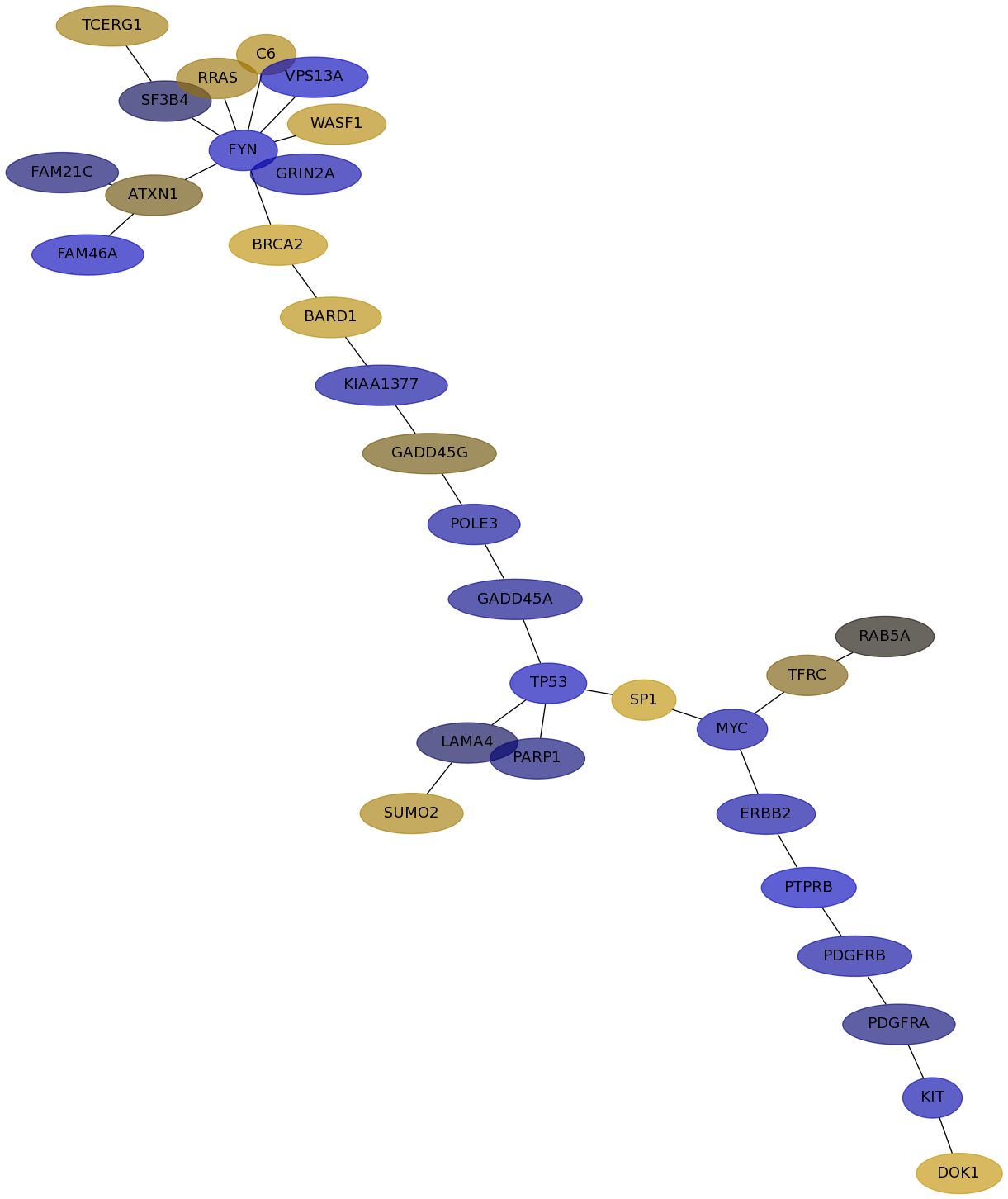
Score for each gene in subnetwork 6832 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| RRAS |   | 1 | 1195 | 1351 | 1384 | 0.086 | 0.036 | -0.035 | 0.016 | -0.010 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| WASF1 |   | 7 | 256 | 1 | 47 | 0.162 | -0.070 | 0.348 | -0.037 | 0.130 |
|---|
| C6 |   | 4 | 440 | 913 | 904 | 0.128 | 0.055 | 0.169 | 0.161 | 0.054 |
|---|
| DOK1 |   | 1 | 1195 | 1351 | 1384 | 0.235 | -0.069 | 0.137 | -0.030 | -0.056 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| RAB5A |   | 2 | 743 | 1351 | 1344 | 0.002 | 0.087 | 0.154 | -0.093 | 0.045 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| VPS13A |   | 10 | 167 | 236 | 234 | -0.203 | 0.105 | -0.046 | 0.120 | 0.062 |
|---|
| FAM21C |   | 1 | 1195 | 1351 | 1384 | -0.028 | -0.042 | 0.158 | -0.090 | -0.062 |
|---|
| PDGFRA |   | 10 | 167 | 141 | 146 | -0.035 | 0.008 | 0.118 | 0.107 | 0.119 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| TCERG1 |   | 4 | 440 | 412 | 419 | 0.098 | 0.093 | 0.172 | 0.228 | 0.105 |
|---|
| TFRC |   | 5 | 360 | 957 | 938 | 0.038 | -0.066 | 0.076 | -0.108 | -0.014 |
|---|
| FYN |   | 10 | 167 | 1 | 40 | -0.171 | -0.167 | 0.073 | -0.137 | 0.110 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| GADD45A |   | 11 | 148 | 1351 | 1308 | -0.057 | 0.076 | -0.099 | 0.109 | 0.093 |
|---|
| PTPRB |   | 5 | 360 | 991 | 977 | -0.199 | 0.006 | 0.149 | 0.094 | 0.131 |
|---|
| SF3B4 |   | 2 | 743 | 1351 | 1344 | -0.017 | 0.017 | -0.199 | 0.167 | -0.007 |
|---|
| FAM46A |   | 2 | 743 | 1351 | 1344 | -0.177 | 0.090 | undef | 0.125 | undef |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| GRIN2A |   | 12 | 140 | 179 | 192 | -0.113 | -0.068 | 0.162 | 0.125 | undef |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| PDGFRB |   | 12 | 140 | 141 | 142 | -0.094 | 0.224 | 0.127 | 0.153 | 0.189 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
GO Enrichment output for subnetwork 6832 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 1.303E-06 | 3.182E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 1.303E-06 | 1.591E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 2.788E-06 | 2.271E-03 |
|---|
| response to X-ray | GO:0010165 |  | 3.089E-06 | 1.887E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.304E-06 | 1.614E-03 |
|---|
| pigmentation | GO:0043473 |  | 3.612E-06 | 1.471E-03 |
|---|
| response to gamma radiation | GO:0010332 |  | 3.925E-06 | 1.37E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.925E-06 | 1.199E-03 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 7.298E-06 | 1.981E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 7.993E-06 | 1.953E-03 |
|---|
| centrosome cycle | GO:0007098 |  | 8.743E-06 | 1.942E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 1.426E-06 | 3.431E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 1.426E-06 | 1.716E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.203E-06 | 2.568E-03 |
|---|
| response to X-ray | GO:0010165 |  | 3.382E-06 | 2.034E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.71E-06 | 1.785E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 3.721E-06 | 1.492E-03 |
|---|
| pigmentation | GO:0043473 |  | 4.056E-06 | 1.394E-03 |
|---|
| response to gamma radiation | GO:0010332 |  | 4.297E-06 | 1.292E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 4.297E-06 | 1.149E-03 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 7.988E-06 | 1.922E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 8.97E-06 | 1.962E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.3E-06 | 5.29E-03 |
|---|
| response to X-ray | GO:0010165 |  | 2.3E-06 | 2.645E-03 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 2.3E-06 | 1.763E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 2.3E-06 | 1.322E-03 |
|---|
| pigmentation | GO:0043473 |  | 3.447E-06 | 1.586E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.505E-06 | 1.344E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 3.714E-06 | 1.22E-03 |
|---|
| response to gamma radiation | GO:0010332 |  | 4.2E-06 | 1.208E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.251E-06 | 1.086E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 6.861E-06 | 1.578E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 6.923E-06 | 1.447E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.206E-06 | 4.065E-03 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.091E-06 | 2.848E-03 |
|---|
| regulation of cell migration | GO:0030334 |  | 3.704E-06 | 2.276E-03 |
|---|
| regulation of locomotion | GO:0040012 |  | 6.294E-06 | 2.9E-03 |
|---|
| regulation of cell motion | GO:0051270 |  | 6.294E-06 | 2.32E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.173E-05 | 6.676E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 2.457E-05 | 6.468E-03 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 3.485E-05 | 8.028E-03 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 4.09E-05 | 8.376E-03 |
|---|
| response to radiation | GO:0009314 |  | 7.992E-05 | 0.01472867 |
|---|
| memory | GO:0007613 |  | 1.159E-04 | 0.01941678 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.3E-06 | 5.29E-03 |
|---|
| response to X-ray | GO:0010165 |  | 2.3E-06 | 2.645E-03 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 2.3E-06 | 1.763E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 2.3E-06 | 1.322E-03 |
|---|
| pigmentation | GO:0043473 |  | 3.447E-06 | 1.586E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.505E-06 | 1.344E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 3.714E-06 | 1.22E-03 |
|---|
| response to gamma radiation | GO:0010332 |  | 4.2E-06 | 1.208E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.251E-06 | 1.086E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 6.861E-06 | 1.578E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 6.923E-06 | 1.447E-03 |
|---|



