Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6815
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7499 | 3.774e-03 | 6.270e-04 | 7.805e-01 | 1.847e-06 |
|---|
| Loi | 0.2307 | 7.960e-02 | 1.119e-02 | 4.266e-01 | 3.799e-04 |
|---|
| Schmidt | 0.6756 | 0.000e+00 | 0.000e+00 | 4.262e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.428e-03 | 0.000e+00 |
|---|
| Wang | 0.2635 | 2.433e-03 | 4.153e-02 | 3.855e-01 | 3.895e-05 |
|---|
Expression data for subnetwork 6815 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
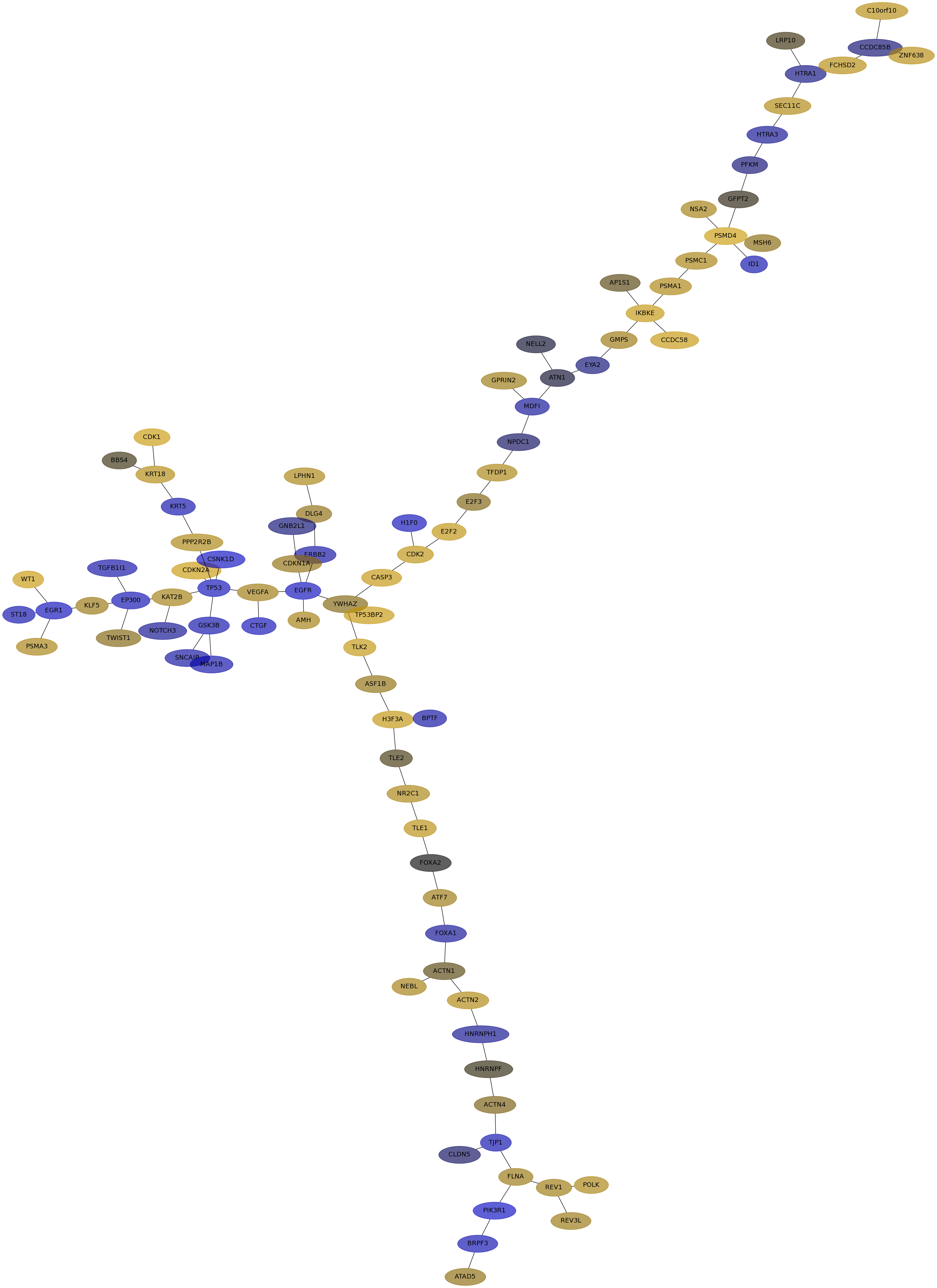
Score for each gene in subnetwork 6815 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| GFPT2 |   | 1 | 1195 | 806 | 838 | 0.004 | 0.096 | -0.142 | undef | 0.176 |
|---|
| CASP3 |   | 11 | 148 | 141 | 143 | 0.234 | -0.032 | -0.142 | -0.097 | -0.084 |
|---|
| CLDN5 |   | 2 | 743 | 806 | 806 | -0.019 | 0.030 | 0.223 | -0.002 | 0.037 |
|---|
| PSMA3 |   | 1 | 1195 | 806 | 838 | 0.116 | -0.149 | -0.008 | -0.003 | 0.014 |
|---|
| ID1 |   | 3 | 557 | 806 | 792 | -0.128 | -0.030 | 0.124 | 0.069 | -0.142 |
|---|
| CCDC58 |   | 1 | 1195 | 806 | 838 | 0.241 | -0.030 | undef | undef | undef |
|---|
| C10orf10 |   | 1 | 1195 | 806 | 838 | 0.160 | 0.075 | 0.075 | 0.040 | 0.019 |
|---|
| HTRA1 |   | 7 | 256 | 83 | 102 | -0.046 | 0.184 | 0.051 | 0.132 | 0.224 |
|---|
| BPTF |   | 4 | 440 | 318 | 337 | -0.098 | 0.159 | 0.132 | undef | 0.030 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| NELL2 |   | 7 | 256 | 727 | 714 | -0.005 | 0.138 | 0.421 | -0.027 | 0.074 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| BRPF3 |   | 2 | 743 | 806 | 806 | -0.145 | 0.028 | undef | 0.218 | undef |
|---|
| LPHN1 |   | 1 | 1195 | 806 | 838 | 0.121 | 0.052 | 0.249 | -0.177 | 0.089 |
|---|
| TLK2 |   | 1 | 1195 | 806 | 838 | 0.231 | 0.044 | 0.106 | undef | 0.107 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| NOTCH3 |   | 5 | 360 | 806 | 784 | -0.063 | 0.115 | 0.136 | undef | -0.082 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| NR2C1 |   | 8 | 222 | 727 | 703 | 0.121 | 0.039 | 0.119 | -0.197 | -0.026 |
|---|
| ZNF638 |   | 5 | 360 | 806 | 784 | 0.164 | 0.088 | 0.020 | 0.077 | -0.045 |
|---|
| KRT5 |   | 10 | 167 | 806 | 780 | -0.125 | 0.046 | 0.064 | -0.006 | -0.190 |
|---|
| E2F3 |   | 2 | 743 | 806 | 806 | 0.041 | -0.091 | 0.177 | -0.082 | 0.050 |
|---|
| HNRNPF |   | 2 | 743 | 806 | 806 | 0.005 | -0.112 | 0.124 | 0.001 | -0.055 |
|---|
| DLG4 |   | 5 | 360 | 806 | 784 | 0.059 | -0.113 | 0.151 | -0.131 | 0.123 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| ASF1B |   | 1 | 1195 | 806 | 838 | 0.062 | -0.265 | 0.031 | 0.019 | -0.174 |
|---|
| NSA2 |   | 4 | 440 | 806 | 790 | 0.103 | 0.097 | 0.132 | -0.026 | 0.006 |
|---|
| FOXA2 |   | 2 | 743 | 806 | 806 | 0.000 | -0.024 | -0.078 | undef | 0.153 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| TLE1 |   | 4 | 440 | 727 | 723 | 0.183 | 0.050 | 0.030 | 0.080 | 0.055 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| AP1S1 |   | 1 | 1195 | 806 | 838 | 0.015 | -0.097 | -0.000 | 0.092 | -0.033 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| H3F3A |   | 2 | 743 | 318 | 346 | 0.221 | 0.232 | 0.151 | 0.237 | -0.142 |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| PSMC1 |   | 1 | 1195 | 806 | 838 | 0.115 | -0.086 | 0.135 | -0.047 | -0.123 |
|---|
| EYA2 |   | 13 | 124 | 318 | 316 | -0.034 | 0.113 | -0.142 | undef | -0.019 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| CSNK1D |   | 6 | 301 | 614 | 602 | -0.222 | -0.044 | 0.170 | -0.088 | 0.013 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| BBS4 |   | 1 | 1195 | 806 | 838 | 0.007 | 0.089 | -0.133 | undef | -0.068 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| TJP1 |   | 8 | 222 | 318 | 325 | -0.139 | 0.102 | 0.099 | 0.319 | 0.049 |
|---|
| ACTN2 |   | 2 | 743 | 806 | 806 | 0.139 | -0.106 | 0.155 | undef | 0.127 |
|---|
| HNRNPH1 |   | 9 | 196 | 614 | 597 | -0.059 | 0.248 | 0.125 | 0.144 | -0.133 |
|---|
| FCHSD2 |   | 2 | 743 | 806 | 806 | 0.165 | -0.228 | 0.098 | -0.145 | 0.047 |
|---|
| NEBL |   | 1 | 1195 | 806 | 838 | 0.104 | 0.046 | -0.046 | -0.053 | -0.185 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| AMH |   | 10 | 167 | 366 | 362 | 0.103 | 0.056 | 0.113 | -0.187 | 0.301 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| GMPS |   | 7 | 256 | 318 | 328 | 0.083 | -0.022 | 0.044 | -0.019 | 0.151 |
|---|
| HTRA3 |   | 1 | 1195 | 806 | 838 | -0.063 | 0.089 | undef | undef | undef |
|---|
| REV1 |   | 9 | 196 | 296 | 285 | 0.090 | -0.027 | 0.165 | 0.049 | 0.039 |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| TLE2 |   | 4 | 440 | 806 | 790 | 0.010 | 0.052 | 0.202 | undef | 0.093 |
|---|
| POLK |   | 3 | 557 | 806 | 792 | 0.120 | 0.046 | undef | 0.113 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SEC11C |   | 2 | 743 | 806 | 806 | 0.141 | -0.215 | undef | undef | undef |
|---|
| PSMA1 |   | 2 | 743 | 806 | 806 | 0.129 | -0.118 | 0.125 | undef | -0.082 |
|---|
| GNB2L1 |   | 1 | 1195 | 806 | 838 | -0.033 | 0.124 | -0.019 | -0.085 | 0.066 |
|---|
| PSMD4 |   | 11 | 148 | 296 | 282 | 0.285 | -0.243 | -0.043 | 0.087 | -0.045 |
|---|
| NPDC1 |   | 3 | 557 | 727 | 728 | -0.019 | 0.221 | 0.044 | 0.006 | -0.146 |
|---|
| PFKM |   | 6 | 301 | 83 | 105 | -0.030 | -0.079 | 0.293 | 0.023 | 0.118 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| REV3L |   | 2 | 743 | 806 | 806 | 0.080 | 0.041 | 0.146 | -0.076 | 0.257 |
|---|
| ATAD5 |   | 1 | 1195 | 806 | 838 | 0.058 | -0.121 | -0.088 | undef | 0.051 |
|---|
| IKBKE |   | 10 | 167 | 552 | 539 | 0.227 | -0.041 | -0.095 | -0.197 | 0.079 |
|---|
| H1F0 |   | 1 | 1195 | 806 | 838 | -0.164 | -0.007 | -0.024 | 0.025 | 0.004 |
|---|
| TFDP1 |   | 6 | 301 | 806 | 783 | 0.123 | -0.178 | 0.129 | undef | -0.065 |
|---|
| CCDC85B |   | 3 | 557 | 806 | 792 | -0.030 | -0.293 | 0.123 | undef | 0.115 |
|---|
| LRP10 |   | 1 | 1195 | 806 | 838 | 0.007 | 0.213 | -0.033 | undef | 0.000 |
|---|
| ATN1 |   | 7 | 256 | 412 | 414 | -0.005 | -0.093 | 0.005 | -0.185 | -0.136 |
|---|
| YWHAZ |   | 3 | 557 | 658 | 654 | 0.043 | -0.274 | -0.018 | undef | 0.054 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| KRT18 |   | 8 | 222 | 590 | 569 | 0.148 | 0.100 | -0.083 | 0.024 | -0.053 |
|---|
| SNCAIP |   | 10 | 167 | 366 | 362 | -0.091 | 0.013 | 0.092 | 0.136 | 0.059 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6815 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.747E-09 | 6.71E-06 |
|---|
| regulation of ligase activity | GO:0051340 |  | 4.775E-09 | 5.832E-06 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.647E-08 | 1.341E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.647E-08 | 1.006E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.852E-08 | 9.05E-06 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.607E-08 | 1.061E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.607E-08 | 9.098E-06 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 4.455E-08 | 1.361E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 4.878E-08 | 1.324E-05 |
|---|
| response to UV | GO:0009411 |  | 7.599E-08 | 1.856E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.97E-07 | 4.375E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 5.894E-09 | 1.418E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.022E-08 | 1.229E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 3.255E-08 | 2.611E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.255E-08 | 1.958E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.66E-08 | 1.761E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 5.143E-08 | 2.062E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 5.143E-08 | 1.768E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.593E-08 | 2.284E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 8.766E-08 | 2.344E-05 |
|---|
| response to UV | GO:0009411 |  | 1.065E-07 | 2.562E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 3.844E-07 | 8.409E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.053E-08 | 2.422E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.724E-08 | 1.983E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 5.412E-08 | 4.149E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 5.412E-08 | 3.112E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.169E-08 | 2.838E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 9.008E-08 | 3.453E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 9.008E-08 | 2.96E-05 |
|---|
| response to UV | GO:0009411 |  | 1.317E-07 | 3.787E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.449E-07 | 3.704E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.881E-07 | 4.326E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.743E-07 | 7.825E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 9.088E-08 | 1.675E-04 |
|---|
| response to light stimulus | GO:0009416 |  | 3.373E-06 | 3.108E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.004E-05 | 6.17E-03 |
|---|
| negative regulation of T cell proliferation | GO:0042130 |  | 1.004E-05 | 4.627E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 1.107E-05 | 4.08E-03 |
|---|
| response to radiation | GO:0009314 |  | 1.641E-05 | 5.04E-03 |
|---|
| negative regulation of binding | GO:0051100 |  | 1.731E-05 | 4.559E-03 |
|---|
| negative regulation of lymphocyte proliferation | GO:0050672 |  | 1.747E-05 | 4.025E-03 |
|---|
| negative regulation of T cell activation | GO:0050868 |  | 1.747E-05 | 3.578E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.014E-05 | 3.712E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.54E-05 | 4.256E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.053E-08 | 2.422E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.724E-08 | 1.983E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 5.412E-08 | 4.149E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 5.412E-08 | 3.112E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.169E-08 | 2.838E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 9.008E-08 | 3.453E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 9.008E-08 | 2.96E-05 |
|---|
| response to UV | GO:0009411 |  | 1.317E-07 | 3.787E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.449E-07 | 3.704E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.881E-07 | 4.326E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.743E-07 | 7.825E-05 |
|---|



