Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6780
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7502 | 3.755e-03 | 6.230e-04 | 7.799e-01 | 1.824e-06 |
|---|
| Loi | 0.2308 | 7.954e-02 | 1.117e-02 | 4.264e-01 | 3.788e-04 |
|---|
| Schmidt | 0.6752 | 0.000e+00 | 0.000e+00 | 4.301e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.430e-03 | 0.000e+00 |
|---|
| Wang | 0.2633 | 2.449e-03 | 4.169e-02 | 3.863e-01 | 3.943e-05 |
|---|
Expression data for subnetwork 6780 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
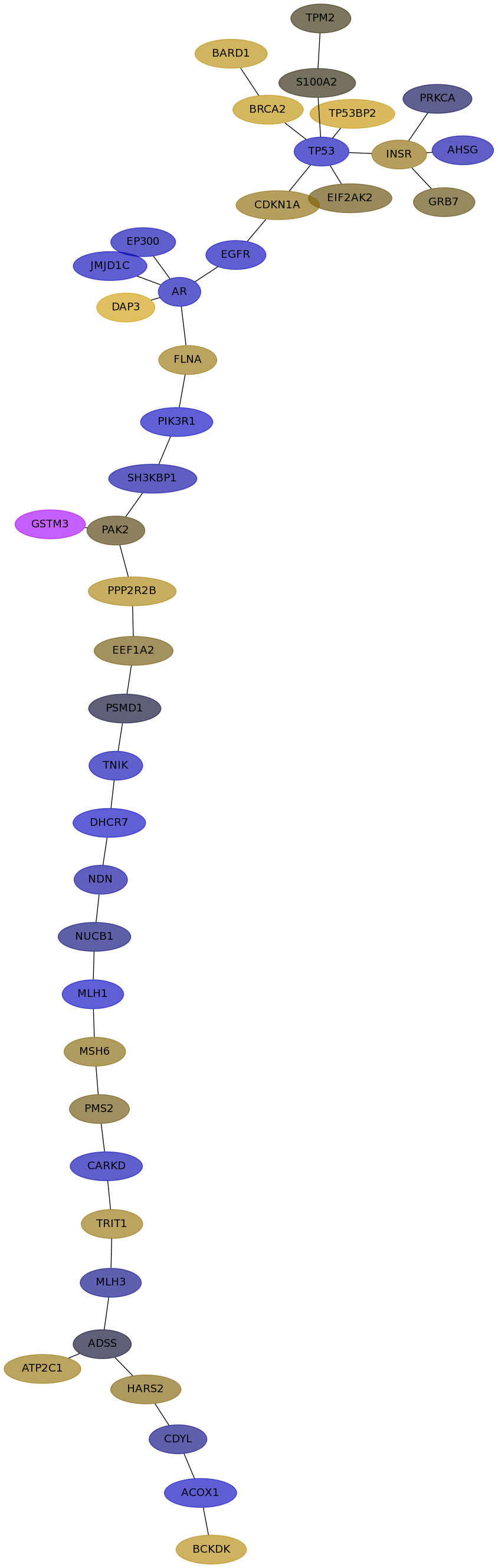
Score for each gene in subnetwork 6780 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TNIK |   | 3 | 557 | 614 | 623 | -0.156 | 0.084 | undef | 0.072 | -0.090 |
|---|
| BCKDK |   | 3 | 557 | 83 | 117 | 0.166 | -0.004 | -0.012 | -0.031 | -0.001 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| PMS2 |   | 2 | 743 | 1425 | 1421 | 0.029 | 0.095 | -0.017 | -0.040 | -0.026 |
|---|
| MLH1 |   | 3 | 557 | 1364 | 1342 | -0.237 | 0.122 | 0.032 | 0.020 | -0.035 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| S100A2 |   | 6 | 301 | 1 | 52 | 0.005 | 0.047 | 0.137 | -0.029 | -0.008 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| MLH3 |   | 2 | 743 | 1425 | 1421 | -0.051 | 0.158 | -0.030 | -0.112 | 0.103 |
|---|
| SH3KBP1 |   | 2 | 743 | 457 | 481 | -0.101 | 0.010 | undef | undef | undef |
|---|
| PSMD1 |   | 1 | 1195 | 1425 | 1445 | -0.006 | -0.059 | 0.001 | 0.123 | -0.008 |
|---|
| EIF2AK2 |   | 9 | 196 | 648 | 629 | 0.023 | -0.022 | -0.134 | 0.096 | -0.303 |
|---|
| EEF1A2 |   | 8 | 222 | 457 | 450 | 0.032 | 0.220 | -0.119 | 0.018 | 0.102 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| CARKD |   | 2 | 743 | 1364 | 1363 | -0.151 | 0.050 | 0.172 | 0.082 | -0.028 |
|---|
| GSTM3 |   | 1 | 1195 | 1425 | 1445 | undef | 0.106 | 0.071 | 0.114 | -0.019 |
|---|
| HARS2 |   | 3 | 557 | 1268 | 1255 | 0.049 | 0.360 | -0.168 | undef | -0.199 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| TPM2 |   | 4 | 440 | 1034 | 1015 | 0.007 | 0.269 | 0.212 | 0.051 | 0.203 |
|---|
| TRIT1 |   | 1 | 1195 | 1425 | 1445 | 0.079 | 0.084 | -0.012 | undef | -0.024 |
|---|
| CDYL |   | 2 | 743 | 1268 | 1259 | -0.050 | 0.116 | -0.203 | undef | 0.206 |
|---|
| JMJD1C |   | 7 | 256 | 552 | 542 | -0.195 | 0.241 | 0.074 | undef | 0.129 |
|---|
| ATP2C1 |   | 2 | 743 | 1010 | 1007 | 0.081 | 0.198 | 0.081 | -0.041 | 0.120 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| ADSS |   | 1 | 1195 | 1425 | 1445 | -0.005 | 0.020 | -0.130 | 0.208 | -0.072 |
|---|
| AHSG |   | 5 | 360 | 412 | 416 | -0.113 | -0.054 | 0.000 | 0.132 | undef |
|---|
| NUCB1 |   | 2 | 743 | 1364 | 1363 | -0.038 | 0.030 | -0.145 | 0.108 | 0.011 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| DAP3 |   | 3 | 557 | 1425 | 1409 | 0.306 | -0.088 | -0.047 | 0.166 | 0.081 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| ACOX1 |   | 9 | 196 | 1 | 44 | -0.209 | 0.122 | -0.104 | 0.252 | -0.094 |
|---|
| NDN |   | 2 | 743 | 648 | 666 | -0.097 | 0.169 | 0.083 | 0.126 | 0.245 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| DHCR7 |   | 1 | 1195 | 1425 | 1445 | -0.232 | -0.033 | -0.089 | 0.239 | 0.106 |
|---|
GO Enrichment output for subnetwork 6780 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.434E-13 | 1.816E-09 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 2.312E-09 | 2.824E-06 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 4.613E-09 | 3.757E-06 |
|---|
| male meiosis | GO:0007140 |  | 4.613E-09 | 2.817E-06 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 9.398E-09 | 4.592E-06 |
|---|
| response to UV | GO:0009411 |  | 1.32E-08 | 5.375E-06 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 1.378E-08 | 4.808E-06 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 5.612E-08 | 1.714E-05 |
|---|
| mismatch repair | GO:0006298 |  | 1.125E-07 | 3.052E-05 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 2.492E-07 | 6.089E-05 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 4.079E-07 | 9.06E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.548E-12 | 3.724E-09 |
|---|
| male meiosis | GO:0007140 |  | 1.447E-09 | 1.741E-06 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 3.368E-09 | 2.701E-06 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 6.719E-09 | 4.041E-06 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.496E-08 | 7.199E-06 |
|---|
| response to UV | GO:0009411 |  | 1.821E-08 | 7.303E-06 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 2.006E-08 | 6.894E-06 |
|---|
| mismatch repair | GO:0006298 |  | 7.659E-08 | 2.304E-05 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 8.913E-08 | 2.383E-05 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 3.622E-07 | 8.713E-05 |
|---|
| somatic diversification of immunoglobulins | GO:0016445 |  | 6.785E-07 | 1.484E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.839E-12 | 8.829E-09 |
|---|
| male meiosis | GO:0007140 |  | 3.942E-09 | 4.533E-06 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 9.169E-09 | 7.03E-06 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 9.169E-09 | 5.272E-06 |
|---|
| response to UV | GO:0009411 |  | 3.245E-08 | 1.493E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.268E-08 | 1.253E-05 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 3.28E-08 | 1.078E-05 |
|---|
| mismatch repair | GO:0006298 |  | 1.132E-07 | 3.253E-05 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 1.9E-07 | 4.857E-05 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 4.636E-07 | 1.066E-04 |
|---|
| somatic diversification of immunoglobulins | GO:0016445 |  | 9.782E-07 | 2.045E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 8.509E-08 | 1.568E-04 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 6.697E-07 | 6.171E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 7.48E-07 | 4.596E-04 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 9.448E-07 | 4.353E-04 |
|---|
| regulation of catabolic process | GO:0009894 |  | 1.256E-06 | 4.631E-04 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 1.336E-06 | 4.103E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.72E-06 | 4.529E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.57E-06 | 8.226E-04 |
|---|
| immunoglobulin production during immune response | GO:0002381 |  | 3.718E-06 | 7.615E-04 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 5.562E-06 | 1.025E-03 |
|---|
| response to UV | GO:0009411 |  | 5.623E-06 | 9.421E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.839E-12 | 8.829E-09 |
|---|
| male meiosis | GO:0007140 |  | 3.942E-09 | 4.533E-06 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 9.169E-09 | 7.03E-06 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 9.169E-09 | 5.272E-06 |
|---|
| response to UV | GO:0009411 |  | 3.245E-08 | 1.493E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.268E-08 | 1.253E-05 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 3.28E-08 | 1.078E-05 |
|---|
| mismatch repair | GO:0006298 |  | 1.132E-07 | 3.253E-05 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 1.9E-07 | 4.857E-05 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 4.636E-07 | 1.066E-04 |
|---|
| somatic diversification of immunoglobulins | GO:0016445 |  | 9.782E-07 | 2.045E-04 |
|---|



