Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6731
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7509 | 3.708e-03 | 6.130e-04 | 7.781e-01 | 1.769e-06 |
|---|
| Loi | 0.2310 | 7.913e-02 | 1.106e-02 | 4.255e-01 | 3.725e-04 |
|---|
| Schmidt | 0.6747 | 0.000e+00 | 0.000e+00 | 4.357e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.438e-03 | 0.000e+00 |
|---|
| Wang | 0.2630 | 2.493e-03 | 4.210e-02 | 3.882e-01 | 4.074e-05 |
|---|
Expression data for subnetwork 6731 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
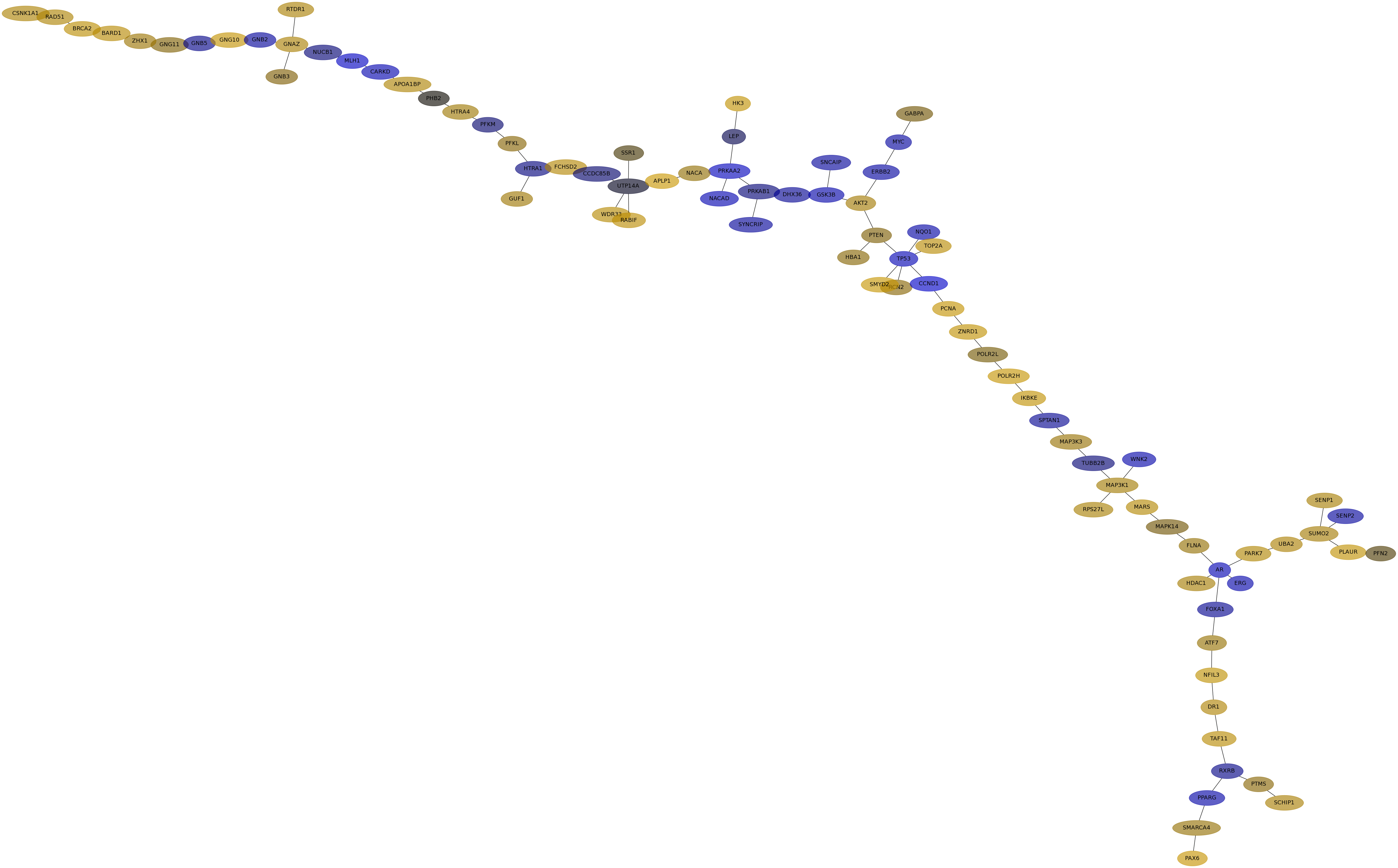
Score for each gene in subnetwork 6731 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| PTMS |   | 2 | 743 | 1294 | 1286 | 0.058 | -0.101 | -0.136 | undef | -0.025 |
|---|
| PRKAB1 |   | 1 | 1195 | 1364 | 1397 | -0.038 | 0.035 | -0.104 | 0.043 | -0.063 |
|---|
| GNB3 |   | 2 | 743 | 1364 | 1363 | 0.046 | -0.115 | -0.028 | 0.013 | -0.119 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| GNG11 |   | 3 | 557 | 1364 | 1342 | 0.049 | -0.039 | 0.135 | 0.004 | 0.106 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| HTRA1 |   | 7 | 256 | 83 | 102 | -0.046 | 0.184 | 0.051 | 0.132 | 0.224 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| SMARCA4 |   | 4 | 440 | 1364 | 1330 | 0.080 | 0.202 | -0.071 | -0.050 | -0.116 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| MAPK14 |   | 4 | 440 | 1 | 59 | 0.033 | -0.065 | 0.299 | 0.015 | 0.119 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| PARK7 |   | 1 | 1195 | 1364 | 1397 | 0.159 | 0.066 | 0.095 | -0.198 | 0.043 |
|---|
| GNB2 |   | 11 | 148 | 525 | 505 | -0.091 | 0.082 | -0.062 | 0.208 | -0.062 |
|---|
| SSR1 |   | 1 | 1195 | 1364 | 1397 | 0.012 | undef | undef | undef | undef |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| UBA2 |   | 1 | 1195 | 1364 | 1397 | 0.137 | -0.103 | 0.102 | -0.101 | 0.113 |
|---|
| GNAZ |   | 15 | 111 | 478 | 457 | 0.134 | -0.116 | 0.308 | 0.267 | 0.011 |
|---|
| TAF11 |   | 3 | 557 | 457 | 463 | 0.192 | 0.112 | 0.237 | 0.053 | 0.180 |
|---|
| ZNRD1 |   | 2 | 743 | 1364 | 1363 | 0.225 | -0.093 | undef | undef | undef |
|---|
| SENP1 |   | 2 | 743 | 1364 | 1363 | 0.126 | -0.034 | undef | -0.075 | undef |
|---|
| MAP3K3 |   | 4 | 440 | 1364 | 1330 | 0.085 | 0.046 | 0.028 | -0.035 | 0.094 |
|---|
| RPS27L |   | 2 | 743 | 1364 | 1363 | 0.131 | 0.039 | -0.033 | -0.098 | 0.026 |
|---|
| POLR2L |   | 5 | 360 | 727 | 722 | 0.037 | -0.012 | 0.224 | 0.054 | 0.015 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| PPARG |   | 7 | 256 | 179 | 201 | -0.119 | 0.011 | 0.105 | 0.059 | 0.039 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| SMYD2 |   | 4 | 440 | 1364 | 1330 | 0.260 | 0.068 | 0.179 | 0.014 | 0.191 |
|---|
| RCN2 |   | 3 | 557 | 412 | 430 | 0.059 | 0.051 | 0.176 | 0.008 | -0.098 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| DR1 |   | 3 | 557 | 1141 | 1129 | 0.151 | 0.041 | 0.174 | -0.054 | 0.104 |
|---|
| GNG10 |   | 3 | 557 | 1190 | 1182 | 0.239 | 0.028 | 0.350 | 0.035 | 0.183 |
|---|
| HTRA4 |   | 8 | 222 | 83 | 101 | 0.102 | undef | undef | undef | undef |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| HDAC1 |   | 4 | 440 | 318 | 337 | 0.122 | -0.020 | 0.050 | -0.035 | -0.172 |
|---|
| POLR2H |   | 2 | 743 | 1364 | 1363 | 0.255 | -0.016 | 0.147 | undef | 0.108 |
|---|
| RTDR1 |   | 5 | 360 | 1364 | 1328 | 0.135 | 0.098 | 0.222 | 0.125 | undef |
|---|
| PRKAA2 |   | 6 | 301 | 1165 | 1138 | -0.204 | 0.148 | 0.086 | 0.250 | -0.078 |
|---|
| PHB2 |   | 2 | 743 | 1364 | 1363 | 0.001 | -0.154 | 0.071 | -0.107 | -0.054 |
|---|
| UTP14A |   | 1 | 1195 | 1364 | 1397 | -0.004 | -0.085 | 0.254 | -0.011 | 0.050 |
|---|
| NACA |   | 5 | 360 | 1165 | 1143 | 0.075 | 0.158 | 0.046 | -0.135 | 0.252 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| MLH1 |   | 3 | 557 | 1364 | 1342 | -0.237 | 0.122 | 0.032 | 0.020 | -0.035 |
|---|
| NACAD |   | 3 | 557 | 1323 | 1309 | -0.159 | 0.096 | 0.093 | 0.155 | -0.078 |
|---|
| PFN2 |   | 11 | 148 | 141 | 143 | 0.014 | -0.001 | 0.286 | -0.002 | -0.056 |
|---|
| CSNK1A1 |   | 2 | 743 | 1364 | 1363 | 0.141 | 0.258 | 0.067 | undef | 0.086 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| HBA1 |   | 3 | 557 | 141 | 170 | 0.058 | -0.042 | 0.140 | 0.142 | 0.131 |
|---|
| APLP1 |   | 8 | 222 | 727 | 703 | 0.284 | 0.043 | -0.105 | -0.121 | -0.190 |
|---|
| FCHSD2 |   | 2 | 743 | 806 | 806 | 0.165 | -0.228 | 0.098 | -0.145 | 0.047 |
|---|
| GNB5 |   | 9 | 196 | 1131 | 1100 | -0.059 | 0.026 | 0.064 | 0.284 | 0.103 |
|---|
| PFKL |   | 7 | 256 | 83 | 102 | 0.056 | 0.106 | -0.173 | 0.160 | -0.158 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| CARKD |   | 2 | 743 | 1364 | 1363 | -0.151 | 0.050 | 0.172 | 0.082 | -0.028 |
|---|
| SCHIP1 |   | 10 | 167 | 478 | 459 | 0.130 | 0.136 | 0.105 | -0.022 | 0.198 |
|---|
| LEP |   | 2 | 743 | 1364 | 1363 | -0.014 | -0.019 | 0.145 | -0.079 | 0.059 |
|---|
| HK3 |   | 2 | 743 | 1364 | 1363 | 0.226 | -0.230 | -0.046 | -0.132 | -0.081 |
|---|
| WNK2 |   | 7 | 256 | 1 | 47 | -0.127 | 0.029 | undef | 0.201 | undef |
|---|
| SPTAN1 |   | 2 | 743 | 1364 | 1363 | -0.074 | 0.035 | 0.077 | 0.067 | -0.025 |
|---|
| MAP3K1 |   | 5 | 360 | 141 | 155 | 0.112 | 0.039 | undef | undef | undef |
|---|
| ERG |   | 9 | 196 | 179 | 195 | -0.134 | -0.026 | 0.235 | -0.027 | 0.182 |
|---|
| PLAUR |   | 5 | 360 | 696 | 680 | 0.232 | 0.083 | 0.022 | -0.030 | 0.108 |
|---|
| RABIF |   | 1 | 1195 | 1364 | 1397 | 0.199 | 0.008 | -0.125 | 0.106 | 0.207 |
|---|
| MARS |   | 2 | 743 | 1364 | 1363 | 0.169 | 0.024 | 0.010 | 0.156 | 0.060 |
|---|
| GUF1 |   | 1 | 1195 | 1364 | 1397 | 0.099 | -0.059 | 0.058 | undef | -0.111 |
|---|
| PFKM |   | 6 | 301 | 83 | 105 | -0.030 | -0.079 | 0.293 | 0.023 | 0.118 |
|---|
| SENP2 |   | 2 | 743 | 696 | 717 | -0.094 | -0.039 | 0.138 | undef | -0.083 |
|---|
| RXRB |   | 1 | 1195 | 1364 | 1397 | -0.058 | 0.194 | -0.015 | undef | 0.137 |
|---|
| NUCB1 |   | 2 | 743 | 1364 | 1363 | -0.038 | 0.030 | -0.145 | 0.108 | 0.011 |
|---|
| SYNCRIP |   | 2 | 743 | 1364 | 1363 | -0.087 | -0.018 | 0.037 | 0.037 | 0.119 |
|---|
| IKBKE |   | 10 | 167 | 552 | 539 | 0.227 | -0.041 | -0.095 | -0.197 | 0.079 |
|---|
| ZHX1 |   | 5 | 360 | 1364 | 1328 | 0.094 | -0.187 | undef | undef | undef |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| NFIL3 |   | 3 | 557 | 1141 | 1129 | 0.222 | -0.163 | -0.095 | -0.029 | 0.130 |
|---|
| CCDC85B |   | 3 | 557 | 806 | 792 | -0.030 | -0.293 | 0.123 | undef | 0.115 |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| APOA1BP |   | 1 | 1195 | 1364 | 1397 | 0.145 | -0.068 | undef | undef | undef |
|---|
| WDR33 |   | 2 | 743 | 1364 | 1363 | 0.174 | -0.117 | 0.001 | -0.067 | 0.083 |
|---|
| DHX36 |   | 1 | 1195 | 1364 | 1397 | -0.078 | 0.073 | undef | undef | undef |
|---|
| SNCAIP |   | 10 | 167 | 366 | 362 | -0.091 | 0.013 | 0.092 | 0.136 | 0.059 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6731 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 5.781E-09 | 1.412E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.726E-08 | 2.109E-05 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 4.01E-08 | 3.265E-05 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 7.982E-08 | 4.875E-05 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 2.373E-07 | 1.159E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 2.373E-07 | 9.661E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.588E-07 | 9.031E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.235E-07 | 9.878E-05 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 3.711E-07 | 1.007E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.804E-06 | 4.407E-04 |
|---|
| endothelial cell migration | GO:0043542 |  | 2.603E-06 | 5.782E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 7.965E-09 | 1.916E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.378E-08 | 2.86E-05 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 5.52E-08 | 4.427E-05 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.098E-07 | 6.607E-05 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 3.263E-07 | 1.57E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 3.263E-07 | 1.308E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.112E-07 | 1.413E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 4.788E-07 | 1.44E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 5.101E-07 | 1.364E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.036E-06 | 7.305E-04 |
|---|
| endothelial cell migration | GO:0043542 |  | 3.57E-06 | 7.81E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.578E-07 | 1.743E-03 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 8.201E-07 | 9.431E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 9.515E-07 | 7.295E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 5.076E-06 | 2.919E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 5.585E-06 | 2.569E-03 |
|---|
| meiosis | GO:0007126 |  | 9.54E-06 | 3.657E-03 |
|---|
| hormone-mediated signaling | GO:0009755 |  | 9.799E-06 | 3.22E-03 |
|---|
| prostate gland development | GO:0030850 |  | 1.009E-05 | 2.901E-03 |
|---|
| male meiosis | GO:0007140 |  | 1.009E-05 | 2.579E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.009E-05 | 2.321E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.009E-05 | 2.11E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| hormone-mediated signaling | GO:0009755 |  | 7.058E-07 | 1.301E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.611E-06 | 1.485E-03 |
|---|
| prostate gland development | GO:0030850 |  | 5.05E-06 | 3.103E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.004E-05 | 4.627E-03 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 1.004E-05 | 3.702E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.107E-05 | 3.4E-03 |
|---|
| response to hormone stimulus | GO:0009725 |  | 1.436E-05 | 3.782E-03 |
|---|
| oxygen transport | GO:0015671 |  | 1.747E-05 | 4.025E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.78E-05 | 5.692E-03 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 2.78E-05 | 5.123E-03 |
|---|
| gas transport | GO:0015669 |  | 4.145E-05 | 6.945E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.578E-07 | 1.743E-03 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 8.201E-07 | 9.431E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 9.515E-07 | 7.295E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 5.076E-06 | 2.919E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 5.585E-06 | 2.569E-03 |
|---|
| meiosis | GO:0007126 |  | 9.54E-06 | 3.657E-03 |
|---|
| hormone-mediated signaling | GO:0009755 |  | 9.799E-06 | 3.22E-03 |
|---|
| prostate gland development | GO:0030850 |  | 1.009E-05 | 2.901E-03 |
|---|
| male meiosis | GO:0007140 |  | 1.009E-05 | 2.579E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.009E-05 | 2.321E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.009E-05 | 2.11E-03 |
|---|



