Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6721
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7511 | 3.690e-03 | 6.090e-04 | 7.775e-01 | 1.747e-06 |
|---|
| Loi | 0.2312 | 7.896e-02 | 1.102e-02 | 4.250e-01 | 3.697e-04 |
|---|
| Schmidt | 0.6746 | 0.000e+00 | 0.000e+00 | 4.367e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.440e-03 | 0.000e+00 |
|---|
| Wang | 0.2629 | 2.503e-03 | 4.220e-02 | 3.887e-01 | 4.106e-05 |
|---|
Expression data for subnetwork 6721 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
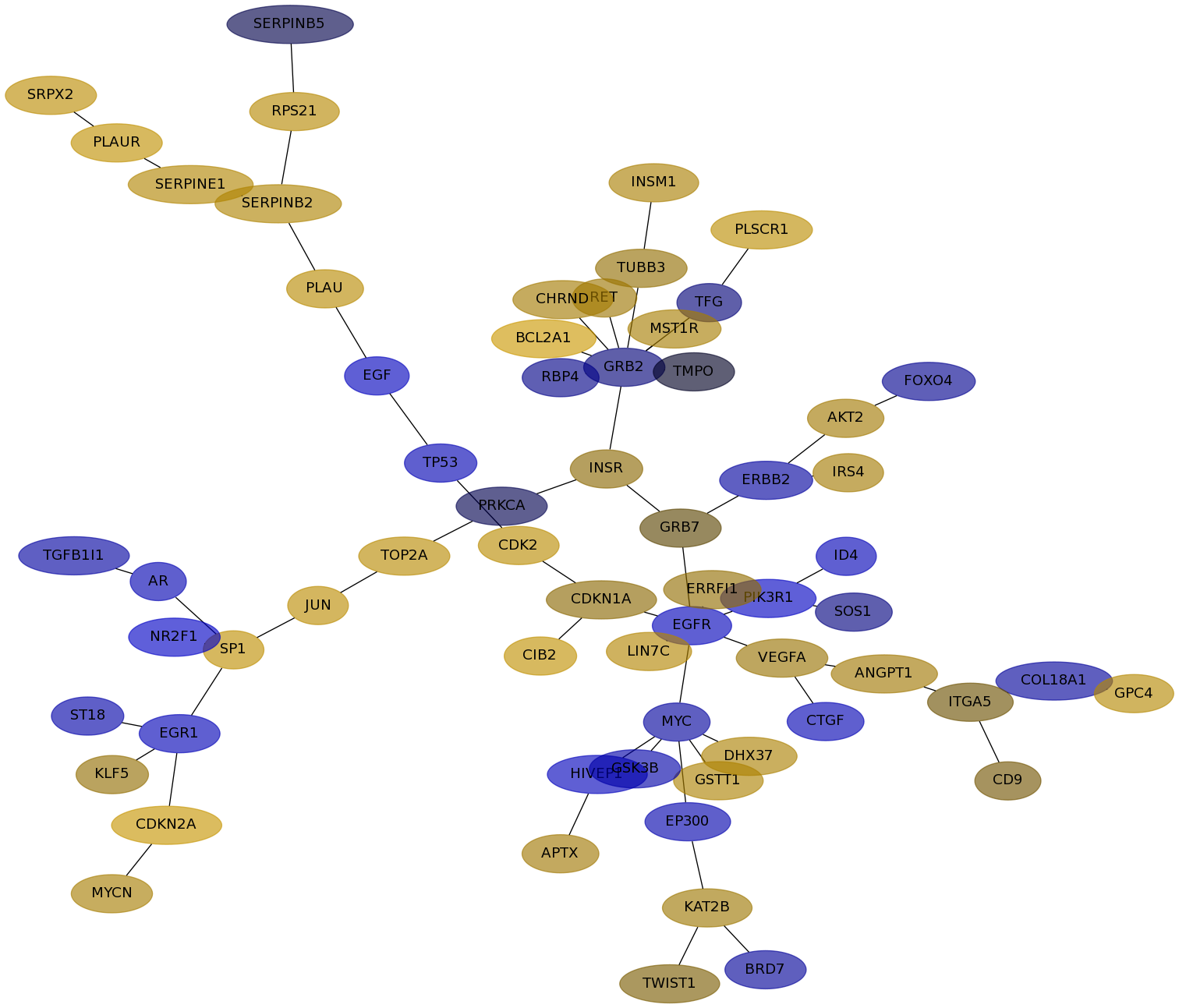
Score for each gene in subnetwork 6721 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| EGF |   | 12 | 140 | 638 | 620 | -0.212 | 0.182 | -0.276 | -0.060 | 0.005 |
|---|
| RET |   | 7 | 256 | 366 | 376 | 0.098 | 0.063 | -0.179 | undef | -0.159 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| COL18A1 |   | 1 | 1195 | 764 | 795 | -0.092 | 0.253 | -0.078 | undef | 0.074 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| PLAU |   | 12 | 140 | 179 | 192 | 0.187 | 0.153 | 0.141 | 0.072 | 0.192 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| RPS21 |   | 1 | 1195 | 764 | 795 | 0.183 | -0.130 | 0.285 | -0.037 | -0.083 |
|---|
| TFG |   | 1 | 1195 | 764 | 795 | -0.040 | 0.155 | -0.191 | 0.045 | 0.238 |
|---|
| TMPO |   | 1 | 1195 | 764 | 795 | -0.005 | -0.236 | 0.054 | 0.027 | 0.122 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| RBP4 |   | 4 | 440 | 764 | 761 | -0.059 | -0.011 | 0.206 | 0.334 | -0.045 |
|---|
| MST1R |   | 5 | 360 | 236 | 242 | 0.124 | 0.083 | -0.212 | -0.001 | 0.026 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| SERPINE1 |   | 8 | 222 | 179 | 200 | 0.160 | 0.200 | 0.056 | 0.070 | 0.030 |
|---|
| ANGPT1 |   | 5 | 360 | 696 | 680 | 0.106 | -0.167 | 0.084 | -0.035 | 0.250 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| HIVEP1 |   | 6 | 301 | 764 | 758 | -0.199 | 0.069 | 0.000 | 0.193 | -0.042 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| CHRND |   | 3 | 557 | 318 | 341 | 0.114 | -0.031 | -0.047 | 0.154 | -0.031 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| SRPX2 |   | 1 | 1195 | 764 | 795 | 0.202 | 0.196 | 0.033 | 0.129 | 0.295 |
|---|
| SERPINB2 |   | 2 | 743 | 764 | 779 | 0.152 | -0.030 | 0.034 | -0.168 | -0.096 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| DHX37 |   | 1 | 1195 | 764 | 795 | 0.141 | -0.085 | undef | undef | undef |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| MYCN |   | 7 | 256 | 764 | 756 | 0.125 | 0.057 | -0.003 | -0.081 | -0.084 |
|---|
| ERRFI1 |   | 10 | 167 | 412 | 408 | 0.076 | 0.215 | undef | -0.043 | undef |
|---|
| TUBB3 |   | 7 | 256 | 764 | 756 | 0.078 | 0.130 | 0.073 | 0.207 | -0.079 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| FOXO4 |   | 1 | 1195 | 764 | 795 | -0.071 | -0.128 | -0.048 | -0.145 | -0.116 |
|---|
| PLSCR1 |   | 10 | 167 | 412 | 408 | 0.202 | -0.159 | 0.091 | undef | -0.105 |
|---|
| SOS1 |   | 5 | 360 | 478 | 476 | -0.049 | 0.002 | -0.007 | 0.160 | -0.042 |
|---|
| PLAUR |   | 5 | 360 | 696 | 680 | 0.232 | 0.083 | 0.022 | -0.030 | 0.108 |
|---|
| GPC4 |   | 1 | 1195 | 764 | 795 | 0.173 | 0.232 | -0.025 | 0.187 | 0.062 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| APTX |   | 1 | 1195 | 764 | 795 | 0.106 | -0.085 | 0.058 | undef | 0.004 |
|---|
| LIN7C |   | 10 | 167 | 590 | 566 | 0.166 | 0.088 | 0.118 | -0.018 | 0.094 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| BCL2A1 |   | 4 | 440 | 764 | 761 | 0.298 | -0.120 | -0.087 | -0.199 | -0.043 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| SERPINB5 |   | 1 | 1195 | 764 | 795 | -0.014 | 0.062 | 0.092 | undef | 0.011 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6721 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.457E-10 | 1.577E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.097E-09 | 1.34E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.632E-08 | 2.958E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.776E-08 | 2.306E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.376E-08 | 4.581E-05 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.138E-07 | 4.632E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.469E-07 | 5.127E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.078E-07 | 6.345E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 2.951E-07 | 8.011E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 5.616E-07 | 1.372E-04 |
|---|
| single strand break repair | GO:0000012 |  | 7.321E-07 | 1.626E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.09E-09 | 2.622E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.85E-09 | 2.226E-06 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 5.377E-08 | 4.313E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.09E-08 | 3.663E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.185E-07 | 5.703E-05 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.449E-07 | 5.811E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.09E-07 | 7.183E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.018E-07 | 9.077E-05 |
|---|
| interphase | GO:0051325 |  | 3.736E-07 | 9.987E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 4.196E-07 | 1.01E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 4.289E-07 | 9.382E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 4.233E-09 | 9.735E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 7.353E-09 | 8.455E-06 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 9.111E-08 | 6.985E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.479E-07 | 8.506E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.029E-07 | 1.393E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.235E-07 | 1.623E-04 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.642E-07 | 1.525E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 9.09E-07 | 2.613E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 9.534E-07 | 2.436E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.534E-07 | 2.193E-04 |
|---|
| interphase | GO:0051325 |  | 1.115E-06 | 2.332E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| wound healing | GO:0042060 |  | 2.363E-07 | 4.354E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.887E-06 | 1.739E-03 |
|---|
| coagulation | GO:0050817 |  | 2.756E-06 | 1.693E-03 |
|---|
| hemostasis | GO:0007599 |  | 4.149E-06 | 1.912E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.172E-06 | 1.538E-03 |
|---|
| ER overload response | GO:0006983 |  | 4.817E-06 | 1.48E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 8.047E-06 | 2.119E-03 |
|---|
| negative regulation of blood coagulation | GO:0030195 |  | 1.337E-05 | 3.08E-03 |
|---|
| regulation of body fluid levels | GO:0050878 |  | 1.761E-05 | 3.607E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.964E-05 | 3.619E-03 |
|---|
| negative regulation of multicellular organismal process | GO:0051241 |  | 2.123E-05 | 3.557E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 4.233E-09 | 9.735E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 7.353E-09 | 8.455E-06 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 9.111E-08 | 6.985E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.479E-07 | 8.506E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.029E-07 | 1.393E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.235E-07 | 1.623E-04 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.642E-07 | 1.525E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 9.09E-07 | 2.613E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 9.534E-07 | 2.436E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.534E-07 | 2.193E-04 |
|---|
| interphase | GO:0051325 |  | 1.115E-06 | 2.332E-04 |
|---|



