Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6679
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7520 | 3.632e-03 | 5.970e-04 | 7.753e-01 | 1.681e-06 |
|---|
| Loi | 0.2309 | 7.927e-02 | 1.110e-02 | 4.258e-01 | 3.746e-04 |
|---|
| Schmidt | 0.6742 | 0.000e+00 | 0.000e+00 | 4.407e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.462e-03 | 0.000e+00 |
|---|
| Wang | 0.2627 | 2.534e-03 | 4.248e-02 | 3.901e-01 | 4.199e-05 |
|---|
Expression data for subnetwork 6679 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
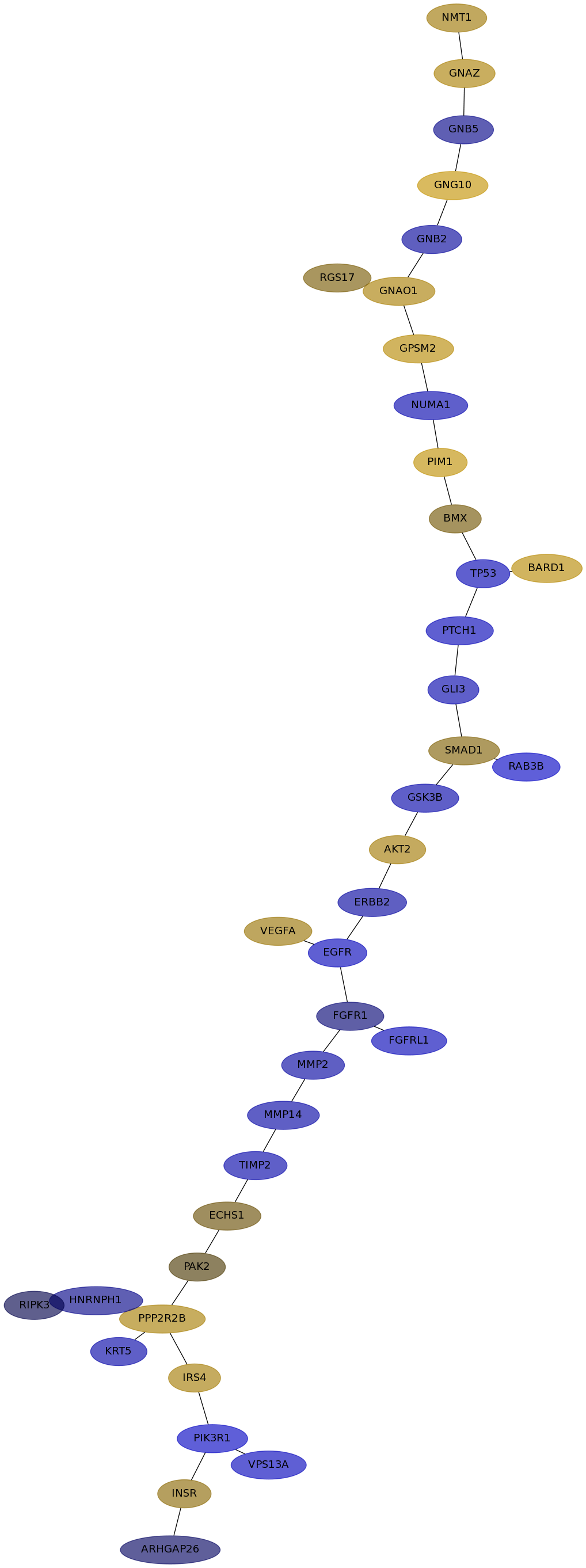
Score for each gene in subnetwork 6679 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| NMT1 |   | 2 | 743 | 1190 | 1189 | 0.099 | -0.161 | -0.183 | -0.057 | 0.077 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| BMX |   | 1 | 1195 | 1190 | 1219 | 0.035 | -0.086 | 0.219 | -0.178 | 0.136 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| HNRNPH1 |   | 9 | 196 | 614 | 597 | -0.059 | 0.248 | 0.125 | 0.144 | -0.133 |
|---|
| GPSM2 |   | 1 | 1195 | 1190 | 1219 | 0.177 | -0.114 | 0.128 | 0.003 | 0.193 |
|---|
| VPS13A |   | 10 | 167 | 236 | 234 | -0.203 | 0.105 | -0.046 | 0.120 | 0.062 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| GNB2 |   | 11 | 148 | 525 | 505 | -0.091 | 0.082 | -0.062 | 0.208 | -0.062 |
|---|
| RGS17 |   | 2 | 743 | 478 | 490 | 0.040 | 0.066 | -0.090 | 0.302 | 0.011 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| GNB5 |   | 9 | 196 | 1131 | 1100 | -0.059 | 0.026 | 0.064 | 0.284 | 0.103 |
|---|
| GNAZ |   | 15 | 111 | 478 | 457 | 0.134 | -0.116 | 0.308 | 0.267 | 0.011 |
|---|
| MMP2 |   | 3 | 557 | 1190 | 1182 | -0.105 | 0.186 | -0.024 | undef | 0.162 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| MMP14 |   | 2 | 743 | 1190 | 1189 | -0.114 | 0.056 | 0.011 | 0.105 | 0.148 |
|---|
| PIM1 |   | 1 | 1195 | 1190 | 1219 | 0.219 | -0.173 | 0.068 | -0.062 | 0.180 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| KRT5 |   | 10 | 167 | 806 | 780 | -0.125 | 0.046 | 0.064 | -0.006 | -0.190 |
|---|
| TIMP2 |   | 2 | 743 | 1190 | 1189 | -0.126 | 0.212 | 0.021 | 0.065 | 0.137 |
|---|
| RAB3B |   | 2 | 743 | 1190 | 1189 | -0.249 | -0.034 | -0.132 | 0.125 | 0.116 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| ECHS1 |   | 2 | 743 | 1190 | 1189 | 0.028 | 0.053 | 0.156 | 0.205 | -0.042 |
|---|
| PTCH1 |   | 5 | 360 | 1190 | 1157 | -0.184 | 0.047 | 0.121 | -0.055 | 0.004 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| NUMA1 |   | 1 | 1195 | 1190 | 1219 | -0.147 | 0.096 | -0.030 | 0.201 | 0.091 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GNAO1 |   | 3 | 557 | 478 | 487 | 0.129 | 0.025 | -0.095 | undef | 0.173 |
|---|
| GNG10 |   | 3 | 557 | 1190 | 1182 | 0.239 | 0.028 | 0.350 | 0.035 | 0.183 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| SMAD1 |   | 5 | 360 | 682 | 671 | 0.049 | 0.159 | -0.062 | 0.140 | 0.048 |
|---|
| FGFRL1 |   | 1 | 1195 | 1190 | 1219 | -0.180 | 0.099 | undef | undef | undef |
|---|
GO Enrichment output for subnetwork 6679 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.401E-06 | 3.423E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.236E-06 | 2.731E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.236E-06 | 1.821E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.961E-06 | 1.808E-03 |
|---|
| mammary gland development | GO:0030879 |  | 1.133E-05 | 5.536E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.202E-05 | 4.894E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.38E-05 | 4.817E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.823E-05 | 5.566E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.991E-05 | 5.405E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.177E-05 | 7.761E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.416E-05 | 9.807E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.51E-06 | 3.634E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.408E-06 | 2.897E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.41E-06 | 1.933E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.41E-06 | 1.45E-03 |
|---|
| mammary gland development | GO:0030879 |  | 1.125E-05 | 5.415E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.131E-05 | 4.535E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.519E-05 | 5.221E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.67E-05 | 5.023E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.19E-05 | 5.856E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.859E-05 | 6.879E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.756E-05 | 0.01040326 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.758E-06 | 4.043E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.759E-06 | 2.023E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.759E-06 | 1.349E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.893E-06 | 2.814E-03 |
|---|
| mammary gland development | GO:0030879 |  | 1.108E-05 | 5.095E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.818E-05 | 6.969E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.093E-05 | 6.876E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.817E-05 | 8.099E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 3.794E-05 | 9.696E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.77E-05 | 0.01327128 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 5.77E-05 | 0.0120648 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 4.753E-08 | 8.76E-05 |
|---|
| ER overload response | GO:0006983 |  | 9.513E-07 | 8.766E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.921E-06 | 1.18E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.652E-06 | 1.222E-03 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 4.706E-06 | 1.735E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 7.75E-06 | 2.381E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 7.75E-06 | 2.04E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 8.698E-06 | 2.004E-03 |
|---|
| lung development | GO:0030324 |  | 9.727E-06 | 1.992E-03 |
|---|
| respiratory tube development | GO:0030323 |  | 1.205E-05 | 2.221E-03 |
|---|
| respiratory system development | GO:0060541 |  | 1.205E-05 | 2.019E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.758E-06 | 4.043E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.759E-06 | 2.023E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.759E-06 | 1.349E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.893E-06 | 2.814E-03 |
|---|
| mammary gland development | GO:0030879 |  | 1.108E-05 | 5.095E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.818E-05 | 6.969E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.093E-05 | 6.876E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.817E-05 | 8.099E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 3.794E-05 | 9.696E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.77E-05 | 0.01327128 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 5.77E-05 | 0.0120648 |
|---|



