Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6659
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7523 | 3.612e-03 | 5.920e-04 | 7.745e-01 | 1.656e-06 |
|---|
| Loi | 0.2308 | 7.948e-02 | 1.115e-02 | 4.263e-01 | 3.779e-04 |
|---|
| Schmidt | 0.6741 | 0.000e+00 | 0.000e+00 | 4.415e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.477e-03 | 0.000e+00 |
|---|
| Wang | 0.2626 | 2.548e-03 | 4.262e-02 | 3.907e-01 | 4.243e-05 |
|---|
Expression data for subnetwork 6659 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
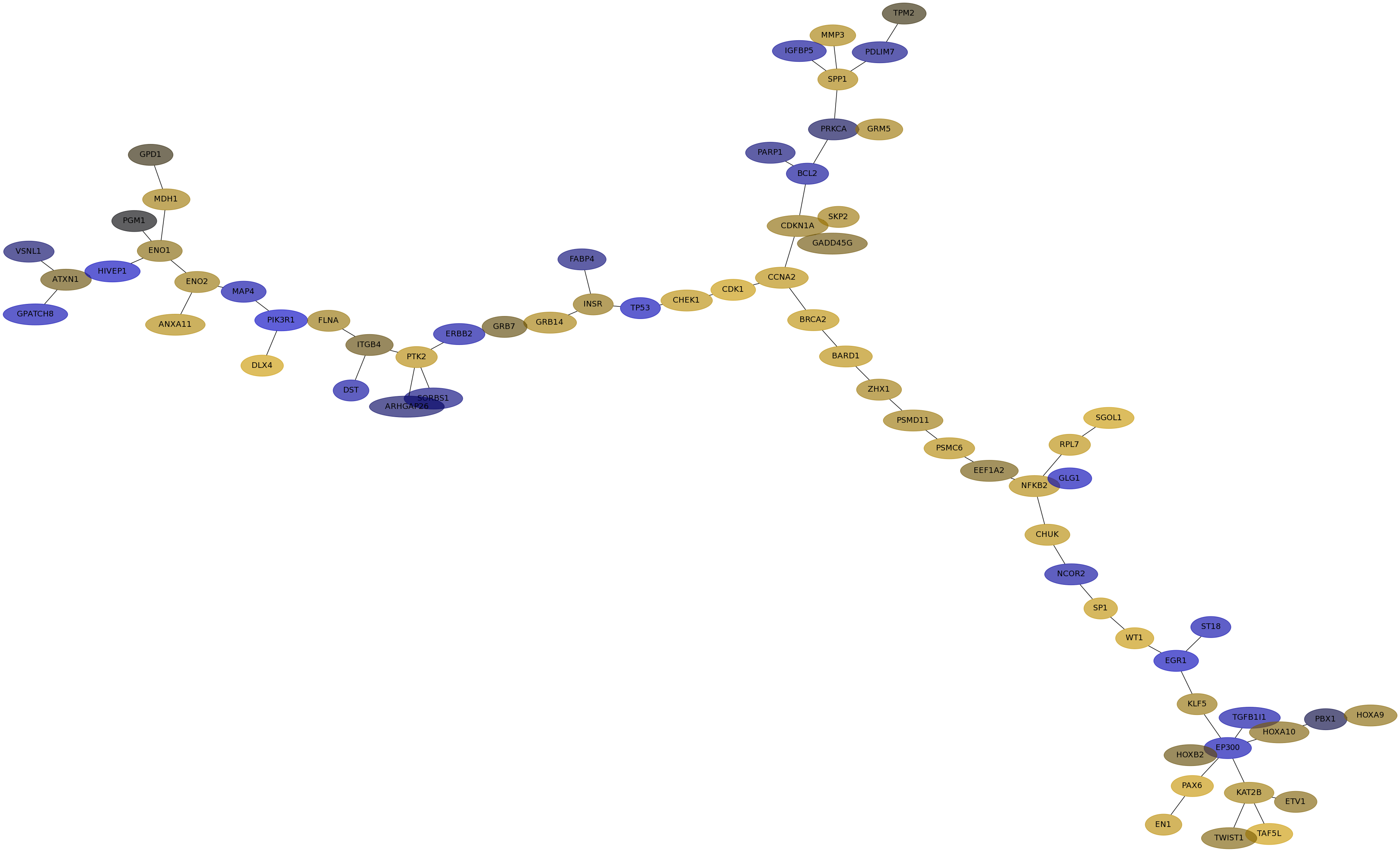
Score for each gene in subnetwork 6659 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PBX1 |   | 6 | 301 | 1 | 52 | -0.010 | 0.145 | -0.013 | 0.176 | -0.053 |
|---|
| CHUK |   | 6 | 301 | 318 | 330 | 0.173 | 0.080 | 0.152 | undef | 0.215 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| GRM5 |   | 15 | 111 | 179 | 191 | 0.094 | -0.019 | 0.000 | 0.093 | -0.093 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| TAF5L |   | 1 | 1195 | 1437 | 1457 | 0.297 | -0.103 | 0.296 | 0.131 | 0.095 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| NFKB2 |   | 7 | 256 | 1116 | 1082 | 0.153 | -0.239 | -0.131 | undef | -0.065 |
|---|
| ENO1 |   | 3 | 557 | 1437 | 1418 | 0.054 | 0.211 | 0.010 | 0.017 | 0.178 |
|---|
| ETV1 |   | 10 | 167 | 1 | 40 | 0.046 | 0.054 | -0.065 | 0.014 | 0.141 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| VSNL1 |   | 3 | 557 | 1437 | 1418 | -0.026 | 0.080 | 0.220 | -0.099 | -0.041 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| MMP3 |   | 6 | 301 | 179 | 205 | 0.121 | 0.171 | 0.081 | 0.198 | 0.192 |
|---|
| PSMC6 |   | 5 | 360 | 318 | 334 | 0.163 | 0.061 | -0.010 | 0.069 | 0.044 |
|---|
| HOXA9 |   | 2 | 743 | 1010 | 1007 | 0.057 | 0.136 | 0.040 | undef | 0.018 |
|---|
| MAP4 |   | 2 | 743 | 1437 | 1433 | -0.115 | 0.217 | 0.221 | 0.086 | 0.268 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| SGOL1 |   | 2 | 743 | 1437 | 1433 | 0.279 | -0.046 | undef | undef | undef |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| HIVEP1 |   | 6 | 301 | 764 | 758 | -0.199 | 0.069 | 0.000 | 0.193 | -0.042 |
|---|
| ANXA11 |   | 1 | 1195 | 1437 | 1457 | 0.152 | 0.087 | 0.067 | undef | 0.006 |
|---|
| FABP4 |   | 6 | 301 | 236 | 239 | -0.036 | -0.028 | 0.227 | 0.072 | 0.107 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| ENO2 |   | 3 | 557 | 727 | 728 | 0.085 | 0.114 | 0.148 | -0.071 | 0.208 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| GPD1 |   | 7 | 256 | 658 | 644 | 0.006 | -0.019 | 0.112 | -0.034 | 0.033 |
|---|
| HOXA10 |   | 4 | 440 | 1 | 59 | 0.046 | 0.203 | -0.057 | -0.160 | 0.016 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| EN1 |   | 10 | 167 | 179 | 194 | 0.196 | 0.024 | 0.011 | -0.016 | 0.102 |
|---|
| EEF1A2 |   | 8 | 222 | 457 | 450 | 0.032 | 0.220 | -0.119 | 0.018 | 0.102 |
|---|
| GPATCH8 |   | 8 | 222 | 614 | 599 | -0.143 | 0.014 | 0.019 | -0.070 | 0.061 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| PSMD11 |   | 6 | 301 | 1437 | 1408 | 0.088 | -0.090 | -0.137 | 0.060 | -0.078 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| DLX4 |   | 10 | 167 | 236 | 234 | 0.291 | 0.057 | -0.004 | undef | -0.059 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PDLIM7 |   | 1 | 1195 | 1437 | 1457 | -0.054 | 0.224 | -0.015 | 0.044 | 0.268 |
|---|
| PGM1 |   | 1 | 1195 | 1437 | 1457 | -0.000 | 0.102 | -0.011 | -0.028 | 0.207 |
|---|
| TPM2 |   | 4 | 440 | 1034 | 1015 | 0.007 | 0.269 | 0.212 | 0.051 | 0.203 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| RPL7 |   | 3 | 557 | 1437 | 1418 | 0.189 | -0.121 | 0.166 | 0.071 | -0.003 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| PTK2 |   | 4 | 440 | 1437 | 1414 | 0.166 | -0.056 | 0.160 | -0.032 | -0.020 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| ZHX1 |   | 5 | 360 | 1364 | 1328 | 0.094 | -0.187 | undef | undef | undef |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| GLG1 |   | 2 | 743 | 1437 | 1433 | -0.165 | 0.090 | -0.094 | 0.050 | -0.091 |
|---|
| MDH1 |   | 2 | 743 | 1437 | 1433 | 0.098 | -0.088 | 0.153 | -0.030 | -0.058 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
GO Enrichment output for subnetwork 6659 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sex determination | GO:0007530 |  | 4.222E-09 | 1.031E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.31E-07 | 2.821E-04 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 7.376E-07 | 6.007E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 1.028E-06 | 6.279E-04 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.607E-06 | 7.851E-04 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 1.714E-06 | 6.978E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 1.852E-06 | 6.464E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.875E-06 | 5.725E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 1.875E-06 | 5.088E-04 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 2.613E-06 | 6.383E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.03E-06 | 6.73E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sex determination | GO:0007530 |  | 4.021E-09 | 9.674E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.907E-07 | 3.497E-04 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 8.649E-07 | 6.936E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 1.205E-06 | 7.249E-04 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 1.536E-06 | 7.391E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 1.636E-06 | 6.559E-04 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.658E-06 | 5.7E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.116E-06 | 6.364E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 2.116E-06 | 5.657E-04 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 2.404E-06 | 5.785E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.193E-06 | 6.984E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sex determination | GO:0007530 |  | 4.53E-09 | 1.042E-05 |
|---|
| reproductive developmental process | GO:0003006 |  | 4.049E-07 | 4.657E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.616E-07 | 4.306E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 1.558E-06 | 8.957E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 3.113E-06 | 1.432E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 4.354E-06 | 1.669E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.772E-06 | 1.568E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.29E-06 | 1.521E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 7.121E-06 | 1.82E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 8.66E-06 | 1.992E-03 |
|---|
| C21-steroid hormone metabolic process | GO:0008207 |  | 1.17E-05 | 2.446E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| hindbrain development | GO:0030902 |  | 4.649E-07 | 8.567E-04 |
|---|
| T cell lineage commitment | GO:0002360 |  | 2.419E-06 | 2.229E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 4.817E-06 | 2.959E-03 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 5.26E-06 | 2.424E-03 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 8.047E-06 | 2.966E-03 |
|---|
| response to gamma radiation | GO:0010332 |  | 8.392E-06 | 2.578E-03 |
|---|
| sex determination | GO:0007530 |  | 1.337E-05 | 3.52E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.337E-05 | 3.08E-03 |
|---|
| T cell differentiation | GO:0030217 |  | 1.409E-05 | 2.885E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.839E-05 | 5.232E-03 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 2.839E-05 | 4.757E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sex determination | GO:0007530 |  | 4.53E-09 | 1.042E-05 |
|---|
| reproductive developmental process | GO:0003006 |  | 4.049E-07 | 4.657E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.616E-07 | 4.306E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 1.558E-06 | 8.957E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 3.113E-06 | 1.432E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 4.354E-06 | 1.669E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.772E-06 | 1.568E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.29E-06 | 1.521E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 7.121E-06 | 1.82E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 8.66E-06 | 1.992E-03 |
|---|
| C21-steroid hormone metabolic process | GO:0008207 |  | 1.17E-05 | 2.446E-03 |
|---|



