Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6657
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7524 | 3.609e-03 | 5.920e-04 | 7.744e-01 | 1.655e-06 |
|---|
| Loi | 0.2307 | 7.961e-02 | 1.119e-02 | 4.266e-01 | 3.800e-04 |
|---|
| Schmidt | 0.6741 | 0.000e+00 | 0.000e+00 | 4.419e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.471e-03 | 0.000e+00 |
|---|
| Wang | 0.2625 | 2.553e-03 | 4.267e-02 | 3.909e-01 | 4.259e-05 |
|---|
Expression data for subnetwork 6657 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
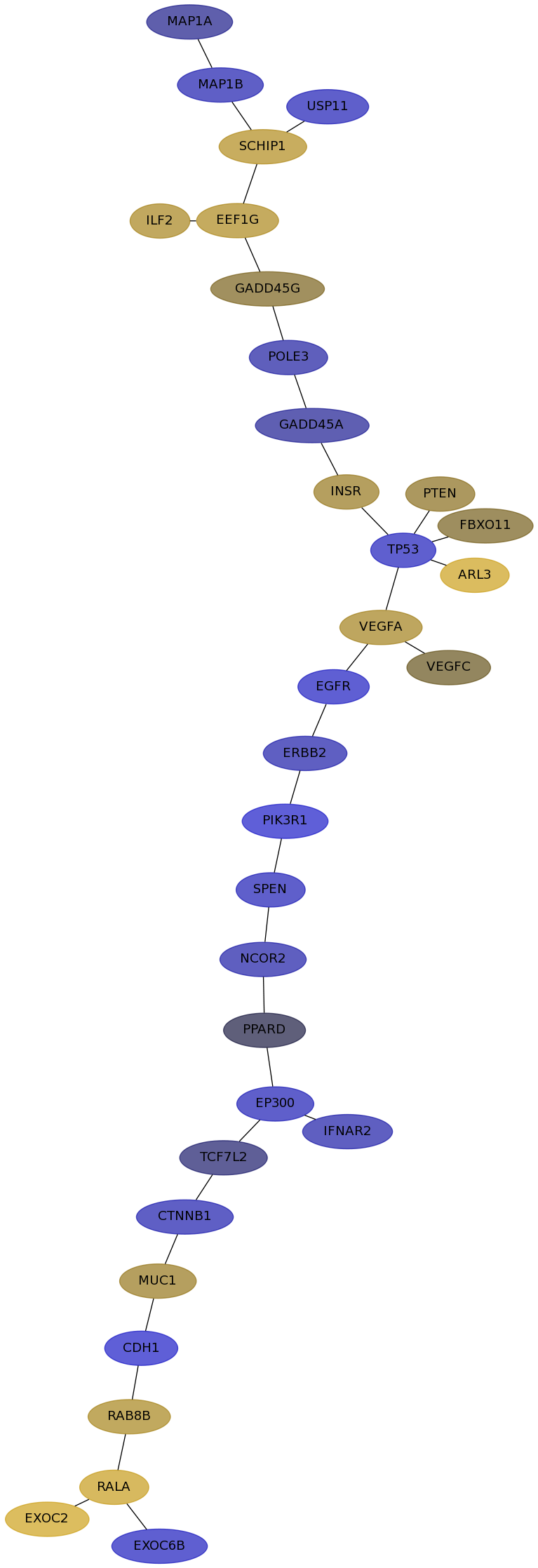
Score for each gene in subnetwork 6657 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| ILF2 |   | 2 | 743 | 1558 | 1545 | 0.099 | -0.123 | -0.068 | 0.116 | 0.004 |
|---|
| VEGFC |   | 2 | 743 | 1558 | 1545 | 0.018 | 0.123 | -0.010 | 0.271 | 0.245 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| IFNAR2 |   | 4 | 440 | 1558 | 1528 | -0.100 | 0.055 | 0.106 | -0.294 | -0.058 |
|---|
| RAB8B |   | 1 | 1195 | 1558 | 1575 | 0.102 | -0.202 | 0.014 | -0.214 | -0.130 |
|---|
| EXOC6B |   | 6 | 301 | 318 | 330 | -0.183 | 0.205 | -0.104 | 0.030 | 0.037 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| EEF1G |   | 8 | 222 | 882 | 869 | 0.116 | -0.224 | 0.131 | 0.086 | 0.082 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| FBXO11 |   | 5 | 360 | 457 | 455 | 0.027 | 0.186 | -0.069 | 0.238 | 0.149 |
|---|
| RALA |   | 10 | 167 | 318 | 323 | 0.224 | 0.207 | -0.047 | 0.169 | 0.146 |
|---|
| ARL3 |   | 6 | 301 | 83 | 105 | 0.266 | -0.002 | 0.203 | 0.206 | -0.029 |
|---|
| PPARD |   | 2 | 743 | 614 | 632 | -0.006 | 0.091 | -0.002 | 0.130 | 0.194 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GADD45A |   | 11 | 148 | 1351 | 1308 | -0.057 | 0.076 | -0.099 | 0.109 | 0.093 |
|---|
| MAP1A |   | 12 | 140 | 1 | 36 | -0.045 | 0.143 | -0.054 | 0.162 | 0.147 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| SPEN |   | 1 | 1195 | 1558 | 1575 | -0.147 | 0.207 | -0.086 | -0.004 | 0.238 |
|---|
| SCHIP1 |   | 10 | 167 | 478 | 459 | 0.130 | 0.136 | 0.105 | -0.022 | 0.198 |
|---|
| EXOC2 |   | 2 | 743 | 1558 | 1545 | 0.279 | 0.060 | -0.105 | 0.067 | -0.098 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| USP11 |   | 7 | 256 | 478 | 462 | -0.144 | 0.106 | 0.121 | 0.218 | -0.044 |
|---|
GO Enrichment output for subnetwork 6657 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.766E-11 | 2.386E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 3.727E-10 | 4.552E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.115E-09 | 9.084E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.29E-09 | 7.876E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 2.597E-09 | 1.269E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.182E-09 | 2.11E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.547E-08 | 5.4E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.955E-08 | 5.971E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.426E-08 | 6.585E-06 |
|---|
| lipid modification | GO:0030258 |  | 2.678E-08 | 6.542E-06 |
|---|
| endothelial cell migration | GO:0043542 |  | 1.726E-07 | 3.833E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.932E-11 | 1.427E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 4.067E-10 | 4.892E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.217E-09 | 9.762E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.432E-09 | 8.615E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 2.833E-09 | 1.363E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.654E-09 | 2.267E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.688E-08 | 5.803E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.205E-08 | 6.63E-06 |
|---|
| lipid modification | GO:0030258 |  | 2.205E-08 | 5.893E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.647E-08 | 6.368E-06 |
|---|
| endothelial cell migration | GO:0043542 |  | 1.882E-07 | 4.117E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.594E-08 | 8.266E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 4.88E-07 | 5.612E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 9.734E-07 | 7.463E-04 |
|---|
| lipid modification | GO:0030258 |  | 1.131E-06 | 6.505E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.251E-06 | 5.753E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.699E-06 | 6.513E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.711E-06 | 8.908E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.711E-06 | 7.794E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 2.711E-06 | 6.928E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 5.779E-06 | 1.329E-03 |
|---|
| inositol metabolic process | GO:0006020 |  | 1.734E-05 | 3.626E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.612E-06 | 6.658E-03 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 4.458E-06 | 4.108E-03 |
|---|
| regulation of cell migration | GO:0030334 |  | 6.868E-06 | 4.22E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.711E-06 | 4.014E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.066E-06 | 3.342E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.017E-05 | 3.125E-03 |
|---|
| regulation of locomotion | GO:0040012 |  | 1.164E-05 | 3.064E-03 |
|---|
| regulation of cell motion | GO:0051270 |  | 1.164E-05 | 2.681E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.409E-05 | 2.885E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.409E-05 | 2.597E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.502E-05 | 2.517E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.594E-08 | 8.266E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 4.88E-07 | 5.612E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 9.734E-07 | 7.463E-04 |
|---|
| lipid modification | GO:0030258 |  | 1.131E-06 | 6.505E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.251E-06 | 5.753E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.699E-06 | 6.513E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.711E-06 | 8.908E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.711E-06 | 7.794E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 2.711E-06 | 6.928E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 5.779E-06 | 1.329E-03 |
|---|
| inositol metabolic process | GO:0006020 |  | 1.734E-05 | 3.626E-03 |
|---|



