Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6641
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7528 | 3.582e-03 | 5.860e-04 | 7.733e-01 | 1.623e-06 |
|---|
| Loi | 0.2306 | 7.989e-02 | 1.126e-02 | 4.273e-01 | 3.844e-04 |
|---|
| Schmidt | 0.6741 | 0.000e+00 | 0.000e+00 | 4.421e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.465e-03 | 0.000e+00 |
|---|
| Wang | 0.2625 | 2.562e-03 | 4.274e-02 | 3.913e-01 | 4.285e-05 |
|---|
Expression data for subnetwork 6641 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
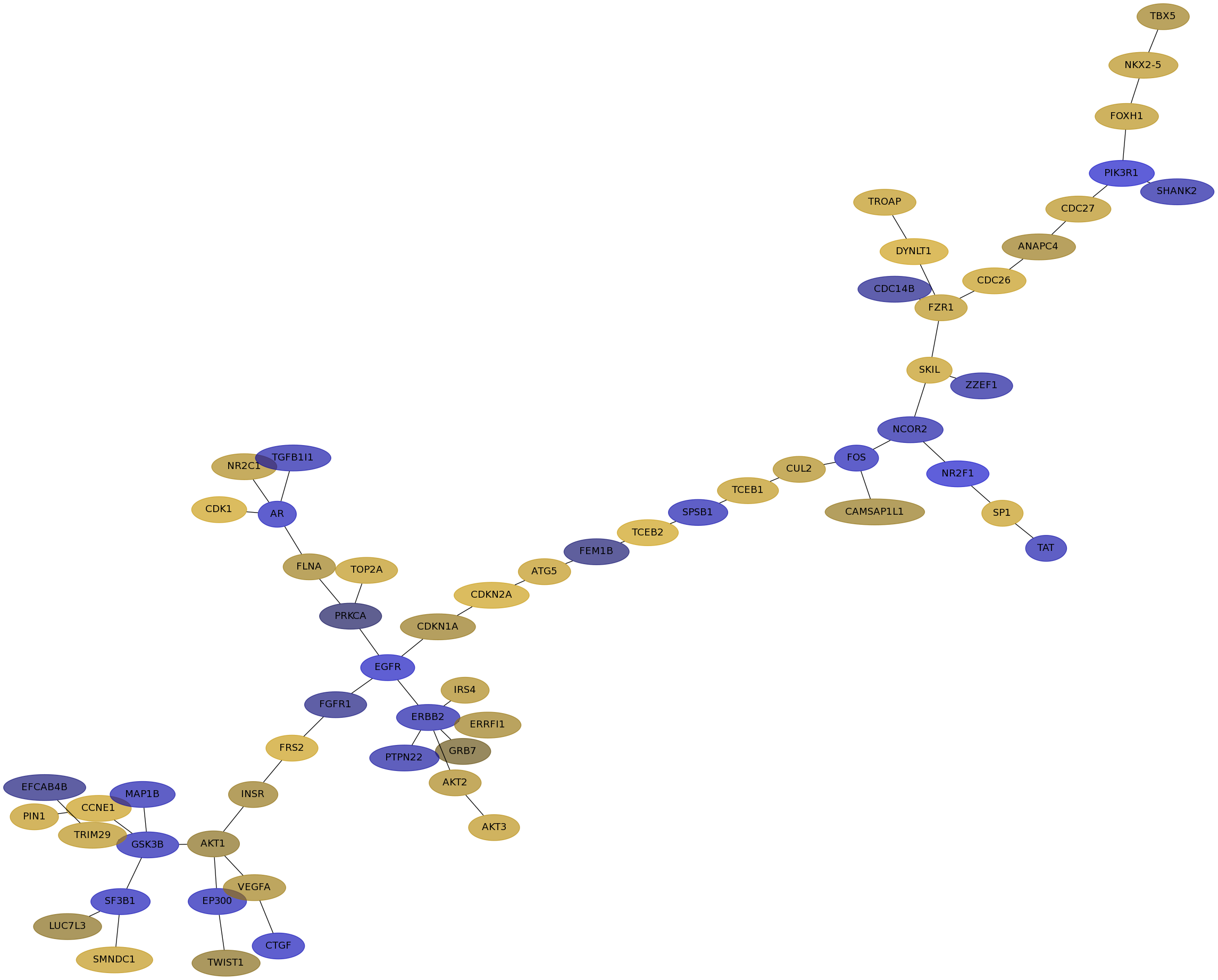
Score for each gene in subnetwork 6641 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TBX5 |   | 1 | 1195 | 1083 | 1111 | 0.078 | 0.020 | 0.085 | 0.042 | 0.188 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| EFCAB4B |   | 1 | 1195 | 1083 | 1111 | -0.032 | 0.047 | undef | undef | undef |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| NKX2-5 |   | 1 | 1195 | 1083 | 1111 | 0.154 | -0.057 | -0.092 | -0.057 | 0.070 |
|---|
| SPSB1 |   | 3 | 557 | 1083 | 1073 | -0.124 | 0.088 | -0.152 | undef | 0.169 |
|---|
| CCNE1 |   | 4 | 440 | 1083 | 1067 | 0.245 | -0.141 | -0.077 | undef | -0.018 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CDC14B |   | 1 | 1195 | 1083 | 1111 | -0.047 | -0.021 | -0.047 | undef | -0.003 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| FZR1 |   | 1 | 1195 | 1083 | 1111 | 0.164 | 0.090 | 0.081 | -0.030 | -0.215 |
|---|
| FEM1B |   | 1 | 1195 | 1083 | 1111 | -0.026 | 0.163 | -0.048 | 0.001 | 0.188 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| ERRFI1 |   | 10 | 167 | 412 | 408 | 0.076 | 0.215 | undef | -0.043 | undef |
|---|
| NR2C1 |   | 8 | 222 | 727 | 703 | 0.121 | 0.039 | 0.119 | -0.197 | -0.026 |
|---|
| FRS2 |   | 2 | 743 | 1083 | 1084 | 0.252 | 0.093 | 0.174 | -0.092 | -0.095 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| TAT |   | 11 | 148 | 83 | 97 | -0.113 | 0.030 | -0.182 | 0.037 | -0.074 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| TCEB2 |   | 2 | 743 | 1083 | 1084 | 0.281 | 0.005 | -0.072 | -0.025 | -0.002 |
|---|
| DYNLT1 |   | 2 | 743 | 727 | 734 | 0.275 | -0.041 | -0.176 | -0.004 | -0.168 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SHANK2 |   | 2 | 743 | 1083 | 1084 | -0.090 | -0.015 | 0.060 | undef | -0.111 |
|---|
| TCEB1 |   | 1 | 1195 | 1083 | 1111 | 0.186 | -0.186 | 0.065 | 0.095 | 0.198 |
|---|
| CAMSAP1L1 |   | 4 | 440 | 296 | 321 | 0.062 | 0.083 | 0.113 | 0.151 | 0.150 |
|---|
| FOXH1 |   | 1 | 1195 | 1083 | 1111 | 0.162 | -0.074 | -0.014 | -0.117 | -0.121 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| PTPN22 |   | 5 | 360 | 957 | 938 | -0.086 | -0.137 | -0.292 | 0.148 | -0.151 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| CDC26 |   | 2 | 743 | 1083 | 1084 | 0.219 | -0.108 | undef | undef | undef |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| TROAP |   | 1 | 1195 | 1083 | 1111 | 0.189 | -0.132 | 0.124 | 0.004 | 0.080 |
|---|
| SKIL |   | 3 | 557 | 780 | 781 | 0.209 | 0.108 | 0.073 | 0.094 | -0.020 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CDC27 |   | 6 | 301 | 1010 | 986 | 0.156 | -0.086 | -0.029 | 0.006 | 0.348 |
|---|
| AKT3 |   | 5 | 360 | 1083 | 1066 | 0.172 | -0.058 | 0.087 | 0.121 | 0.080 |
|---|
| ZZEF1 |   | 1 | 1195 | 1083 | 1111 | -0.074 | 0.151 | 0.010 | -0.170 | 0.210 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| CUL2 |   | 7 | 256 | 842 | 816 | 0.124 | -0.192 | -0.090 | -0.035 | 0.166 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| ATG5 |   | 2 | 743 | 882 | 892 | 0.195 | -0.113 | -0.013 | -0.050 | 0.210 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| ANAPC4 |   | 3 | 557 | 1083 | 1073 | 0.071 | 0.091 | undef | 0.030 | undef |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| SMNDC1 |   | 3 | 557 | 1083 | 1073 | 0.199 | 0.079 | 0.288 | 0.002 | 0.174 |
|---|
| TRIM29 |   | 8 | 222 | 296 | 287 | 0.161 | 0.240 | -0.150 | -0.077 | 0.024 |
|---|
GO Enrichment output for subnetwork 6641 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.879E-09 | 4.59E-06 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 3.585E-09 | 4.379E-06 |
|---|
| regulation of ligase activity | GO:0051340 |  | 5.874E-09 | 4.783E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.007E-08 | 1.226E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 5.54E-08 | 2.707E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.028E-08 | 3.269E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.233E-07 | 4.303E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.153E-07 | 6.575E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 7.88E-07 | 2.139E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 8.597E-07 | 2.1E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.108E-06 | 2.46E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.127E-09 | 9.929E-06 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 8.69E-09 | 1.045E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.421E-08 | 1.139E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.774E-08 | 2.27E-05 |
|---|
| interphase | GO:0051325 |  | 4.822E-08 | 2.32E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.775E-08 | 3.519E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.417E-07 | 4.87E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.174E-07 | 6.539E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 3.405E-07 | 9.104E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.528E-06 | 3.677E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.604E-06 | 3.509E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.674E-08 | 3.851E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.496E-07 | 1.72E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 4.452E-07 | 3.413E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 6.94E-07 | 3.991E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 6.952E-07 | 3.198E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.184E-06 | 8.373E-04 |
|---|
| interphase | GO:0051325 |  | 2.551E-06 | 8.383E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 4.847E-06 | 1.394E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.924E-06 | 2.025E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 9.628E-06 | 2.215E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 9.71E-06 | 2.03E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| heart development | GO:0007507 |  | 6.428E-07 | 1.185E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 5.35E-06 | 4.93E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 5.992E-06 | 3.681E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 6.694E-06 | 3.084E-03 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 6.694E-06 | 2.467E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 8.289E-06 | 2.546E-03 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 8.289E-06 | 2.182E-03 |
|---|
| regulation of ligase activity | GO:0051340 |  | 9.191E-06 | 2.117E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.005E-05 | 4.105E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.136E-05 | 5.78E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.136E-05 | 5.254E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.674E-08 | 3.851E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.496E-07 | 1.72E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 4.452E-07 | 3.413E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 6.94E-07 | 3.991E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 6.952E-07 | 3.198E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.184E-06 | 8.373E-04 |
|---|
| interphase | GO:0051325 |  | 2.551E-06 | 8.383E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 4.847E-06 | 1.394E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.924E-06 | 2.025E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 9.628E-06 | 2.215E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 9.71E-06 | 2.03E-03 |
|---|



