Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6637
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7530 | 3.571e-03 | 5.840e-04 | 7.729e-01 | 1.612e-06 |
|---|
| Loi | 0.2305 | 8.000e-02 | 1.129e-02 | 4.276e-01 | 3.861e-04 |
|---|
| Schmidt | 0.6742 | 0.000e+00 | 0.000e+00 | 4.410e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.465e-03 | 0.000e+00 |
|---|
| Wang | 0.2625 | 2.561e-03 | 4.274e-02 | 3.913e-01 | 4.282e-05 |
|---|
Expression data for subnetwork 6637 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
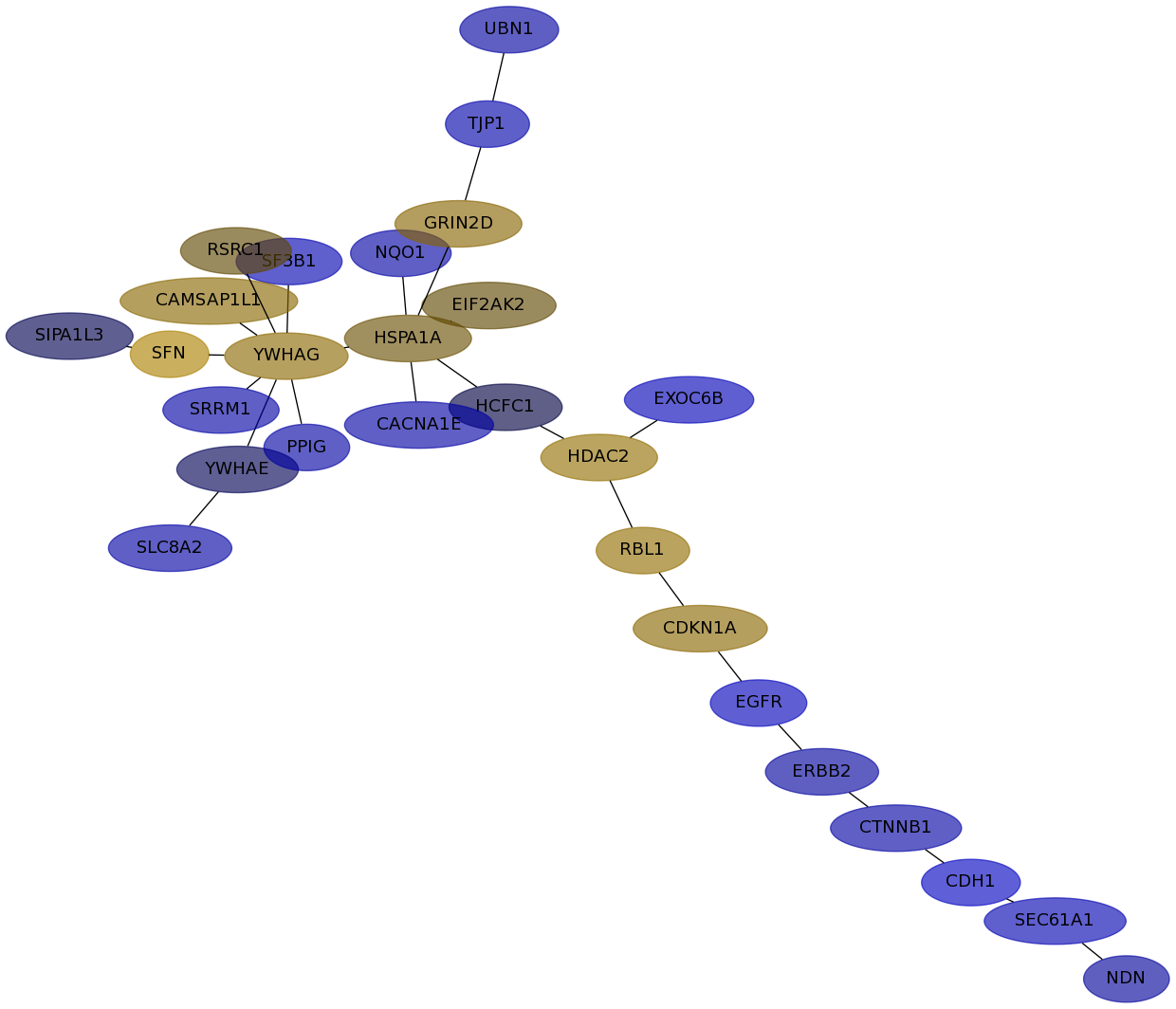
Score for each gene in subnetwork 6637 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| HCFC1 |   | 4 | 440 | 648 | 645 | -0.011 | 0.009 | 0.209 | 0.061 | -0.093 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| UBN1 |   | 3 | 557 | 648 | 647 | -0.106 | 0.059 | 0.130 | -0.144 | 0.066 |
|---|
| CACNA1E |   | 1 | 1195 | 648 | 692 | -0.113 | 0.011 | 0.000 | 0.185 | undef |
|---|
| EXOC6B |   | 6 | 301 | 318 | 330 | -0.183 | 0.205 | -0.104 | 0.030 | 0.037 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| SRRM1 |   | 10 | 167 | 296 | 283 | -0.120 | 0.139 | 0.244 | 0.294 | -0.109 |
|---|
| SLC8A2 |   | 1 | 1195 | 648 | 692 | -0.119 | 0.094 | -0.015 | 0.141 | 0.065 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| TJP1 |   | 8 | 222 | 318 | 325 | -0.139 | 0.102 | 0.099 | 0.319 | 0.049 |
|---|
| CAMSAP1L1 |   | 4 | 440 | 296 | 321 | 0.062 | 0.083 | 0.113 | 0.151 | 0.150 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| YWHAE |   | 4 | 440 | 179 | 209 | -0.018 | 0.112 | 0.279 | -0.094 | 0.014 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| HSPA1A |   | 2 | 743 | 648 | 666 | 0.030 | 0.164 | 0.070 | -0.023 | 0.046 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| SIPA1L3 |   | 3 | 557 | 648 | 647 | -0.016 | 0.039 | -0.062 | 0.136 | 0.049 |
|---|
| EIF2AK2 |   | 9 | 196 | 648 | 629 | 0.023 | -0.022 | -0.134 | 0.096 | -0.303 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| SEC61A1 |   | 1 | 1195 | 648 | 692 | -0.161 | 0.128 | -0.096 | -0.045 | 0.070 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| PPIG |   | 8 | 222 | 648 | 631 | -0.116 | 0.024 | 0.221 | -0.065 | 0.101 |
|---|
| NDN |   | 2 | 743 | 648 | 666 | -0.097 | 0.169 | 0.083 | 0.126 | 0.245 |
|---|
| GRIN2D |   | 13 | 124 | 552 | 522 | 0.058 | 0.155 | 0.000 | undef | undef |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
| HDAC2 |   | 13 | 124 | 318 | 316 | 0.079 | -0.094 | 0.148 | 0.089 | 0.037 |
|---|
GO Enrichment output for subnetwork 6637 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| vesicle docking during exocytosis | GO:0006904 |  | 2.638E-07 | 6.445E-04 |
|---|
| vesicle docking | GO:0048278 |  | 3.602E-07 | 4.4E-04 |
|---|
| membrane docking | GO:0022406 |  | 9.108E-07 | 7.417E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.471E-06 | 8.983E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 4.031E-06 | 1.97E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.634E-06 | 2.701E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.055E-05 | 3.683E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.424E-05 | 4.35E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.508E-05 | 4.094E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.769E-05 | 4.321E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.881E-05 | 4.177E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.087E-06 | 2.614E-03 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 4.089E-06 | 4.919E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.73E-06 | 5.397E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 8.268E-06 | 4.973E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.285E-05 | 6.185E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.363E-05 | 5.467E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.369E-05 | 4.707E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.711E-05 | 5.147E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.298E-05 | 6.143E-03 |
|---|
| regulation of phospholipase A2 activity | GO:0032429 |  | 4.427E-05 | 0.01065168 |
|---|
| glial cell migration | GO:0008347 |  | 4.427E-05 | 9.683E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 8.762E-06 | 0.02015336 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.451E-05 | 0.01668441 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2.193E-05 | 0.01681324 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.141E-05 | 0.02956334 |
|---|
| glial cell migration | GO:0008347 |  | 8.297E-05 | 0.0381684 |
|---|
| regulation of sensory perception | GO:0051931 |  | 8.297E-05 | 0.031807 |
|---|
| axonal fasciculation | GO:0007413 |  | 1.242E-04 | 0.04081834 |
|---|
| neuron migration | GO:0001764 |  | 1.313E-04 | 0.03775271 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.736E-04 | 0.04436363 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.985E-04 | 0.04565433 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.142E-04 | 0.04477702 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.843E-06 | 3.397E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.16E-05 | 0.01068958 |
|---|
| neuron migration | GO:0001764 |  | 4.823E-05 | 0.02962823 |
|---|
| hippocampus development | GO:0021766 |  | 1.063E-04 | 0.04896072 |
|---|
| regulation of phospholipase A2 activity | GO:0032429 |  | 1.063E-04 | 0.03916858 |
|---|
| neuron recognition | GO:0008038 |  | 1.063E-04 | 0.03264048 |
|---|
| cell morphogenesis involved in differentiation | GO:0000904 |  | 1.093E-04 | 0.02877456 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.127E-04 | 0.02596278 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.127E-04 | 0.02307802 |
|---|
| axon extension | GO:0048675 |  | 1.591E-04 | 0.02931666 |
|---|
| limbic system development | GO:0021761 |  | 1.591E-04 | 0.02665151 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 8.762E-06 | 0.02015336 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.451E-05 | 0.01668441 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2.193E-05 | 0.01681324 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.141E-05 | 0.02956334 |
|---|
| glial cell migration | GO:0008347 |  | 8.297E-05 | 0.0381684 |
|---|
| regulation of sensory perception | GO:0051931 |  | 8.297E-05 | 0.031807 |
|---|
| axonal fasciculation | GO:0007413 |  | 1.242E-04 | 0.04081834 |
|---|
| neuron migration | GO:0001764 |  | 1.313E-04 | 0.03775271 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.736E-04 | 0.04436363 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.985E-04 | 0.04565433 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.142E-04 | 0.04477702 |
|---|



