Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6630
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7534 | 3.546e-03 | 5.780e-04 | 7.719e-01 | 1.582e-06 |
|---|
| Loi | 0.2304 | 8.005e-02 | 1.130e-02 | 4.277e-01 | 3.870e-04 |
|---|
| Schmidt | 0.6742 | 0.000e+00 | 0.000e+00 | 4.411e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.463e-03 | 0.000e+00 |
|---|
| Wang | 0.2624 | 2.571e-03 | 4.283e-02 | 3.917e-01 | 4.313e-05 |
|---|
Expression data for subnetwork 6630 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
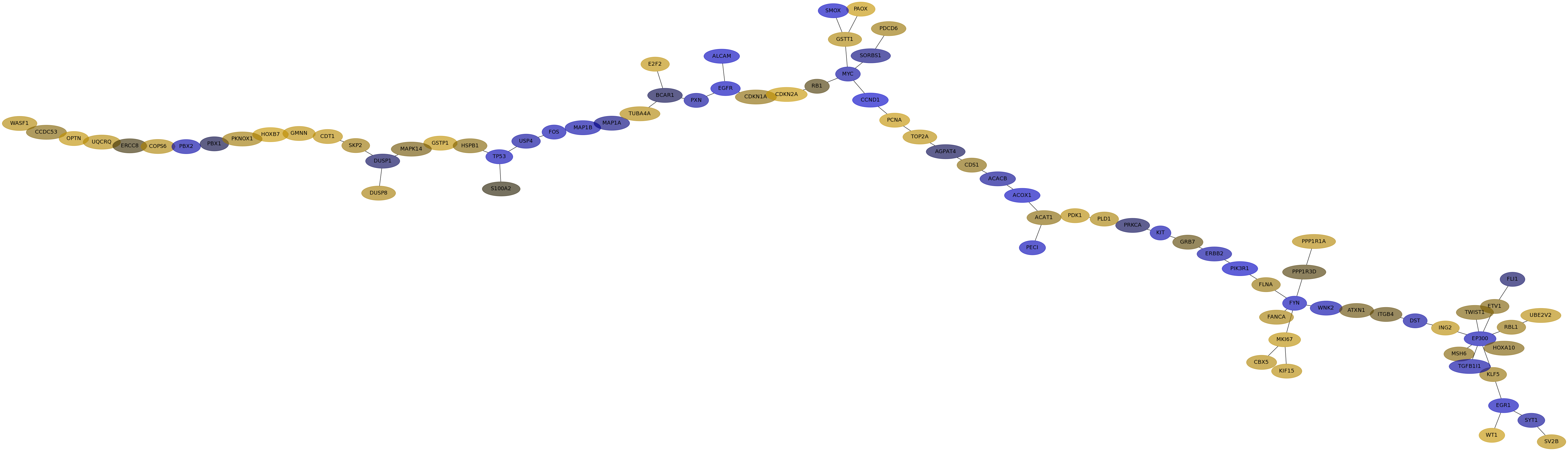
Score for each gene in subnetwork 6630 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ACACB |   | 2 | 743 | 1 | 66 | -0.069 | 0.025 | 0.127 | -0.130 | -0.006 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| HOXB7 |   | 6 | 301 | 1 | 52 | 0.267 | -0.139 | -0.083 | 0.085 | -0.051 |
|---|
| WASF1 |   | 7 | 256 | 1 | 47 | 0.162 | -0.070 | 0.348 | -0.037 | 0.130 |
|---|
| DUSP8 |   | 4 | 440 | 1 | 59 | 0.118 | 0.257 | 0.068 | 0.208 | -0.029 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PBX1 |   | 6 | 301 | 1 | 52 | -0.010 | 0.145 | -0.013 | 0.176 | -0.053 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| MAPK14 |   | 4 | 440 | 1 | 59 | 0.033 | -0.065 | 0.299 | 0.015 | 0.119 |
|---|
| CDS1 |   | 1 | 1195 | 1 | 85 | 0.056 | 0.156 | 0.023 | 0.231 | 0.000 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| ETV1 |   | 10 | 167 | 1 | 40 | 0.046 | 0.054 | -0.065 | 0.014 | 0.141 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| TUBA4A |   | 7 | 256 | 1 | 47 | 0.157 | 0.136 | -0.010 | undef | 0.156 |
|---|
| ERCC8 |   | 1 | 1195 | 1 | 85 | 0.009 | -0.011 | -0.043 | 0.069 | 0.188 |
|---|
| PDK1 |   | 10 | 167 | 1 | 40 | 0.183 | -0.138 | -0.041 | -0.265 | -0.126 |
|---|
| UQCRQ |   | 2 | 743 | 1 | 66 | 0.171 | 0.035 | -0.031 | 0.141 | 0.010 |
|---|
| KIF15 |   | 2 | 743 | 1 | 66 | 0.179 | -0.188 | 0.138 | -0.126 | 0.010 |
|---|
| MKI67 |   | 5 | 360 | 1 | 56 | 0.205 | -0.123 | 0.176 | -0.086 | 0.018 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| ALCAM |   | 8 | 222 | 1 | 46 | -0.208 | 0.193 | 0.067 | 0.088 | -0.010 |
|---|
| PPP1R1A |   | 2 | 743 | 1 | 66 | 0.165 | -0.061 | 0.171 | 0.353 | 0.115 |
|---|
| OPTN |   | 7 | 256 | 1 | 47 | 0.226 | -0.099 | 0.015 | undef | 0.035 |
|---|
| PAOX |   | 3 | 557 | 1 | 63 | 0.255 | -0.190 | 0.163 | undef | -0.086 |
|---|
| FYN |   | 10 | 167 | 1 | 40 | -0.171 | -0.167 | 0.073 | -0.137 | 0.110 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| UBE2V2 |   | 2 | 743 | 1 | 66 | 0.180 | -0.027 | 0.196 | undef | 0.060 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| SV2B |   | 1 | 1195 | 1 | 85 | 0.182 | -0.113 | 0.185 | 0.171 | 0.195 |
|---|
| USP4 |   | 7 | 256 | 1 | 47 | -0.093 | 0.147 | 0.097 | -0.392 | 0.054 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| CCDC53 |   | 1 | 1195 | 1 | 85 | 0.052 | -0.133 | 0.035 | 0.032 | -0.052 |
|---|
| CDT1 |   | 3 | 557 | 1 | 63 | 0.194 | -0.184 | -0.072 | undef | -0.030 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| GSTP1 |   | 14 | 117 | 1 | 34 | 0.222 | -0.166 | 0.201 | 0.126 | -0.136 |
|---|
| S100A2 |   | 6 | 301 | 1 | 52 | 0.005 | 0.047 | 0.137 | -0.029 | -0.008 |
|---|
| HOXA10 |   | 4 | 440 | 1 | 59 | 0.046 | 0.203 | -0.057 | -0.160 | 0.016 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| GMNN |   | 2 | 743 | 1 | 66 | 0.201 | -0.071 | 0.104 | 0.066 | 0.158 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| MAP1A |   | 12 | 140 | 1 | 36 | -0.045 | 0.143 | -0.054 | 0.162 | 0.147 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| SMOX |   | 2 | 743 | 1 | 66 | -0.232 | 0.276 | -0.089 | undef | 0.023 |
|---|
| PECI |   | 2 | 743 | 1 | 66 | -0.174 | 0.127 | 0.056 | -0.011 | 0.051 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| FLI1 |   | 2 | 743 | 1 | 66 | -0.021 | 0.018 | 0.067 | -0.175 | 0.009 |
|---|
| WNK2 |   | 7 | 256 | 1 | 47 | -0.127 | 0.029 | undef | 0.201 | undef |
|---|
| PKNOX1 |   | 1 | 1195 | 1 | 85 | 0.103 | 0.102 | 0.152 | -0.019 | 0.028 |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| DUSP1 |   | 4 | 440 | 1 | 59 | -0.019 | 0.127 | 0.170 | -0.031 | 0.134 |
|---|
| PPP1R3D |   | 1 | 1195 | 1 | 85 | 0.015 | -0.004 | 0.267 | 0.127 | -0.155 |
|---|
| PXN |   | 12 | 140 | 1 | 36 | -0.088 | 0.284 | -0.022 | 0.060 | 0.114 |
|---|
| AGPAT4 |   | 2 | 743 | 1 | 66 | -0.014 | 0.005 | 0.059 | -0.020 | 0.108 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ING2 |   | 3 | 557 | 1 | 63 | 0.189 | 0.168 | 0.051 | 0.026 | -0.054 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| BCAR1 |   | 6 | 301 | 1 | 52 | -0.012 | 0.229 | undef | 0.223 | undef |
|---|
| FANCA |   | 9 | 196 | 1 | 44 | 0.120 | -0.188 | 0.106 | -0.127 | -0.053 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| SYT1 |   | 11 | 148 | 1 | 39 | -0.079 | 0.051 | 0.038 | 0.189 | -0.124 |
|---|
| PBX2 |   | 5 | 360 | 1 | 56 | -0.122 | -0.073 | 0.264 | -0.095 | 0.092 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| PDCD6 |   | 10 | 167 | 1 | 40 | 0.091 | -0.060 | undef | undef | undef |
|---|
| ACOX1 |   | 9 | 196 | 1 | 44 | -0.209 | 0.122 | -0.104 | 0.252 | -0.094 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| ACAT1 |   | 2 | 743 | 1 | 66 | 0.057 | -0.011 | 0.087 | 0.038 | 0.112 |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| CBX5 |   | 5 | 360 | 1 | 56 | 0.155 | 0.016 | 0.148 | 0.030 | -0.009 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6630 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.905E-08 | 4.654E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.234E-08 | 2.729E-05 |
|---|
| response to UV | GO:0009411 |  | 8.052E-08 | 6.557E-05 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 2.916E-07 | 1.781E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.956E-07 | 1.933E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.371E-07 | 1.78E-04 |
|---|
| sex determination | GO:0007530 |  | 4.742E-07 | 1.655E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 4.742E-07 | 1.448E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 6.416E-07 | 1.742E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 7.26E-07 | 1.774E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 3.239E-06 | 7.193E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.082E-08 | 5.008E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.441E-08 | 2.937E-05 |
|---|
| response to UV | GO:0009411 |  | 6.755E-08 | 5.418E-05 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.209E-07 | 1.931E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.519E-07 | 1.693E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.901E-07 | 1.564E-04 |
|---|
| sex determination | GO:0007530 |  | 4.037E-07 | 1.388E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 5.072E-07 | 1.525E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.794E-07 | 1.549E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 7.764E-07 | 1.868E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 2.581E-06 | 5.645E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.375E-08 | 1.006E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.401E-08 | 8.511E-05 |
|---|
| response to UV | GO:0009411 |  | 1.203E-07 | 9.224E-05 |
|---|
| sex determination | GO:0007530 |  | 4.591E-07 | 2.64E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5.493E-07 | 2.527E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 6.202E-07 | 2.377E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 6.708E-07 | 2.204E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 8.808E-07 | 2.532E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.292E-06 | 3.302E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.012E-06 | 4.628E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 4.875E-06 | 1.019E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 2.538E-06 | 4.678E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 9.562E-06 | 8.811E-03 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 1.204E-05 | 7.397E-03 |
|---|
| response to radiation | GO:0009314 |  | 1.326E-05 | 6.108E-03 |
|---|
| response to X-ray | GO:0010165 |  | 1.565E-05 | 5.768E-03 |
|---|
| actin filament organization | GO:0007015 |  | 3.269E-05 | 0.01003986 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.997E-05 | 0.01052251 |
|---|
| response to light stimulus | GO:0009416 |  | 4.844E-05 | 0.0111605 |
|---|
| signal complex assembly | GO:0007172 |  | 7.215E-05 | 0.01477417 |
|---|
| regulation of DNA replication | GO:0006275 |  | 9.116E-05 | 0.01680045 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 1.237E-04 | 0.02072054 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.375E-08 | 1.006E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.401E-08 | 8.511E-05 |
|---|
| response to UV | GO:0009411 |  | 1.203E-07 | 9.224E-05 |
|---|
| sex determination | GO:0007530 |  | 4.591E-07 | 2.64E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5.493E-07 | 2.527E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 6.202E-07 | 2.377E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 6.708E-07 | 2.204E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 8.808E-07 | 2.532E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.292E-06 | 3.302E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.012E-06 | 4.628E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 4.875E-06 | 1.019E-03 |
|---|



