Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6605
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7537 | 3.524e-03 | 5.740e-04 | 7.711e-01 | 1.560e-06 |
|---|
| Loi | 0.2305 | 7.999e-02 | 1.129e-02 | 4.275e-01 | 3.860e-04 |
|---|
| Schmidt | 0.6737 | 0.000e+00 | 0.000e+00 | 4.461e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.462e-03 | 0.000e+00 |
|---|
| Wang | 0.2621 | 2.606e-03 | 4.315e-02 | 3.932e-01 | 4.422e-05 |
|---|
Expression data for subnetwork 6605 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
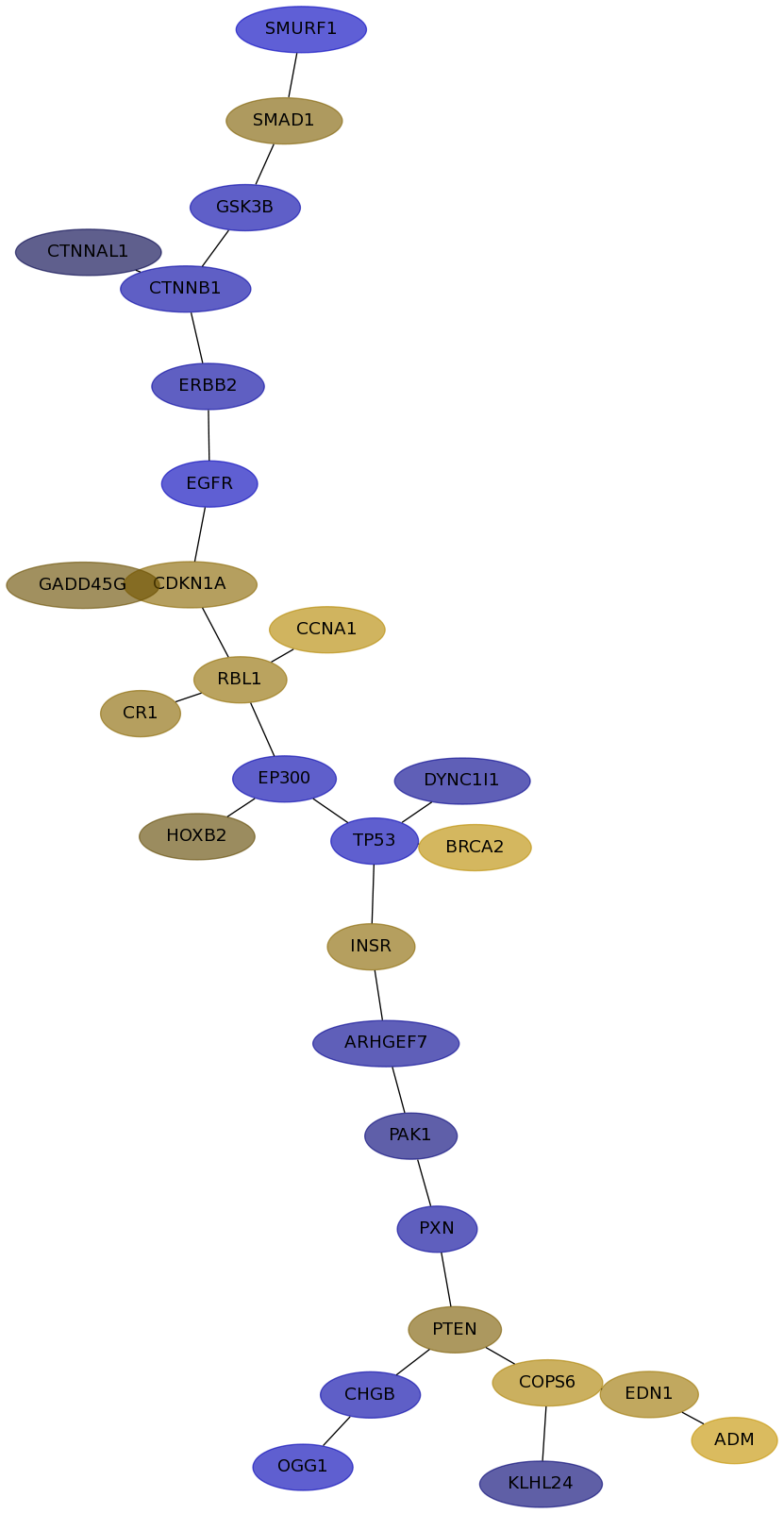
Score for each gene in subnetwork 6605 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| PAK1 |   | 3 | 557 | 682 | 685 | -0.040 | -0.122 | -0.143 | 0.075 | 0.006 |
|---|
| CHGB |   | 7 | 256 | 682 | 664 | -0.126 | -0.041 | 0.018 | 0.102 | -0.131 |
|---|
| KLHL24 |   | 4 | 440 | 682 | 678 | -0.036 | -0.011 | 0.062 | 0.044 | -0.125 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| PXN |   | 12 | 140 | 1 | 36 | -0.088 | 0.284 | -0.022 | 0.060 | 0.114 |
|---|
| CTNNAL1 |   | 4 | 440 | 682 | 678 | -0.014 | 0.073 | -0.008 | -0.131 | 0.004 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| EDN1 |   | 6 | 301 | 682 | 669 | 0.100 | 0.126 | 0.249 | -0.086 | 0.121 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| DYNC1I1 |   | 5 | 360 | 614 | 612 | -0.069 | 0.013 | -0.086 | -0.045 | 0.088 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| ADM |   | 10 | 167 | 682 | 653 | 0.255 | 0.187 | 0.091 | 0.169 | 0.212 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| CR1 |   | 1 | 1195 | 682 | 732 | 0.064 | -0.040 | 0.230 | 0.076 | -0.070 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| SMAD1 |   | 5 | 360 | 682 | 671 | 0.049 | 0.159 | -0.062 | 0.140 | 0.048 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| SMURF1 |   | 5 | 360 | 682 | 671 | -0.222 | 0.236 | -0.049 | undef | -0.006 |
|---|
| OGG1 |   | 1 | 1195 | 682 | 732 | -0.174 | 0.139 | -0.104 | 0.150 | -0.050 |
|---|
GO Enrichment output for subnetwork 6605 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.187E-10 | 2.9E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.556E-10 | 4.343E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 8.282E-10 | 6.745E-07 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.654E-09 | 1.01E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.556E-09 | 1.738E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 4.944E-09 | 2.013E-06 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 7.756E-09 | 2.707E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 7.756E-09 | 2.368E-06 |
|---|
| endothelial cell migration | GO:0043542 |  | 5.536E-08 | 1.503E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 5.536E-08 | 1.353E-05 |
|---|
| cardiac muscle tissue development | GO:0048738 |  | 5.536E-08 | 1.23E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.335E-10 | 3.213E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.999E-10 | 4.811E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 9.314E-10 | 7.47E-07 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.86E-09 | 1.119E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.185E-09 | 2.014E-06 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.559E-09 | 2.229E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 5.559E-09 | 1.911E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 8.721E-09 | 2.623E-06 |
|---|
| endothelial cell migration | GO:0043542 |  | 6.224E-08 | 1.664E-05 |
|---|
| inositol metabolic process | GO:0006020 |  | 6.224E-08 | 1.497E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 6.224E-08 | 1.361E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.011E-07 | 2.326E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.064E-07 | 1.223E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.272E-07 | 9.751E-05 |
|---|
| response to UV | GO:0009411 |  | 1.424E-07 | 8.189E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.814E-07 | 8.346E-05 |
|---|
| purine deoxyribonucleotide catabolic process | GO:0009155 |  | 1.814E-07 | 6.955E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.152E-07 | 7.072E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.416E-07 | 6.947E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.668E-07 | 6.818E-05 |
|---|
| male meiosis | GO:0007140 |  | 3.622E-07 | 8.33E-05 |
|---|
| ER overload response | GO:0006983 |  | 3.622E-07 | 7.573E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 1.085E-08 | 2E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.162E-08 | 1.071E-05 |
|---|
| heart development | GO:0007507 |  | 1.177E-08 | 7.234E-06 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 1.276E-08 | 5.881E-06 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.088E-08 | 1.138E-05 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.699E-08 | 1.136E-05 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 7.269E-08 | 1.914E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 7.976E-08 | 1.838E-05 |
|---|
| base-excision repair | GO:0006284 |  | 1.114E-07 | 2.281E-05 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.516E-07 | 2.793E-05 |
|---|
| purine deoxyribonucleotide metabolic process | GO:0009151 |  | 3.714E-07 | 6.222E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.011E-07 | 2.326E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.064E-07 | 1.223E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.272E-07 | 9.751E-05 |
|---|
| response to UV | GO:0009411 |  | 1.424E-07 | 8.189E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.814E-07 | 8.346E-05 |
|---|
| purine deoxyribonucleotide catabolic process | GO:0009155 |  | 1.814E-07 | 6.955E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.152E-07 | 7.072E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.416E-07 | 6.947E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.668E-07 | 6.818E-05 |
|---|
| male meiosis | GO:0007140 |  | 3.622E-07 | 8.33E-05 |
|---|
| ER overload response | GO:0006983 |  | 3.622E-07 | 7.573E-05 |
|---|



