Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6598
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7538 | 3.518e-03 | 5.730e-04 | 7.708e-01 | 1.554e-06 |
|---|
| Loi | 0.2306 | 7.988e-02 | 1.126e-02 | 4.273e-01 | 3.842e-04 |
|---|
| Schmidt | 0.6736 | 0.000e+00 | 0.000e+00 | 4.471e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.466e-03 | 0.000e+00 |
|---|
| Wang | 0.2620 | 2.614e-03 | 4.323e-02 | 3.936e-01 | 4.448e-05 |
|---|
Expression data for subnetwork 6598 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
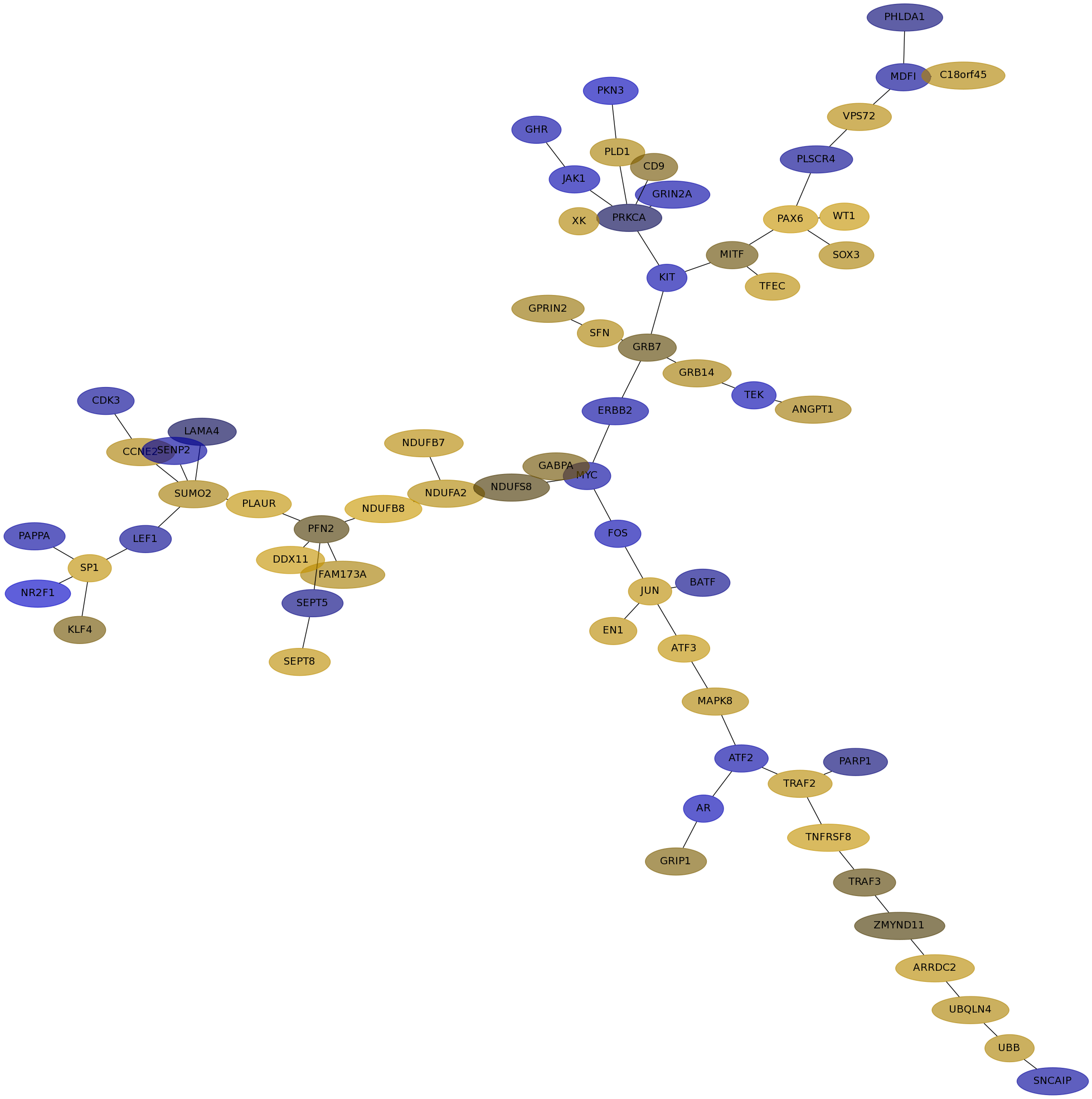
Score for each gene in subnetwork 6598 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| CCNE2 |   | 11 | 148 | 412 | 407 | 0.136 | -0.178 | 0.241 | undef | -0.055 |
|---|
| MAPK8 |   | 8 | 222 | 696 | 677 | 0.153 | 0.200 | 0.249 | -0.066 | 0.138 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| SOX3 |   | 1 | 1195 | 696 | 744 | 0.135 | 0.003 | 0.000 | 0.184 | -0.175 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| TNFRSF8 |   | 1 | 1195 | 696 | 744 | 0.241 | -0.067 | 0.247 | -0.123 | -0.296 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| PHLDA1 |   | 2 | 743 | 696 | 717 | -0.036 | -0.020 | 0.122 | -0.119 | 0.053 |
|---|
| CDK3 |   | 7 | 256 | 590 | 570 | -0.076 | 0.189 | -0.041 | -0.089 | 0.082 |
|---|
| TFEC |   | 1 | 1195 | 696 | 744 | 0.190 | -0.270 | -0.201 | undef | -0.046 |
|---|
| MITF |   | 5 | 360 | 696 | 680 | 0.027 | 0.032 | 0.065 | 0.145 | 0.183 |
|---|
| NDUFB8 |   | 5 | 360 | 696 | 680 | 0.286 | -0.012 | 0.340 | -0.171 | 0.205 |
|---|
| ATF2 |   | 1 | 1195 | 696 | 744 | -0.114 | -0.129 | -0.037 | -0.014 | 0.133 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| UBQLN4 |   | 4 | 440 | 696 | 689 | 0.143 | -0.114 | undef | -0.213 | undef |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| BATF |   | 1 | 1195 | 696 | 744 | -0.056 | -0.144 | -0.269 | -0.087 | -0.147 |
|---|
| FAM173A |   | 1 | 1195 | 696 | 744 | 0.125 | -0.114 | -0.218 | undef | 0.169 |
|---|
| LEF1 |   | 3 | 557 | 696 | 695 | -0.068 | 0.016 | -0.018 | 0.001 | 0.139 |
|---|
| JAK1 |   | 5 | 360 | 696 | 680 | -0.140 | 0.034 | -0.176 | 0.111 | 0.159 |
|---|
| KLF4 |   | 1 | 1195 | 696 | 744 | 0.035 | 0.060 | -0.026 | 0.157 | -0.025 |
|---|
| TRAF3 |   | 1 | 1195 | 696 | 744 | 0.019 | -0.208 | 0.234 | undef | -0.025 |
|---|
| SEPT5 |   | 2 | 743 | 696 | 717 | -0.050 | -0.078 | -0.021 | 0.176 | 0.075 |
|---|
| NDUFB7 |   | 1 | 1195 | 696 | 744 | 0.172 | -0.088 | 0.144 | undef | 0.029 |
|---|
| ANGPT1 |   | 5 | 360 | 696 | 680 | 0.106 | -0.167 | 0.084 | -0.035 | 0.250 |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| DDX11 |   | 3 | 557 | 179 | 213 | 0.254 | -0.126 | -0.191 | -0.033 | -0.135 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| PAPPA |   | 1 | 1195 | 696 | 744 | -0.099 | 0.148 | -0.054 | undef | -0.011 |
|---|
| TRAF2 |   | 3 | 557 | 658 | 654 | 0.185 | -0.172 | -0.013 | undef | -0.092 |
|---|
| PFN2 |   | 11 | 148 | 141 | 143 | 0.014 | -0.001 | 0.286 | -0.002 | -0.056 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| EN1 |   | 10 | 167 | 179 | 194 | 0.196 | 0.024 | 0.011 | -0.016 | 0.102 |
|---|
| NDUFA2 |   | 2 | 743 | 696 | 717 | 0.159 | -0.014 | 0.084 | -0.037 | -0.022 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| VPS72 |   | 6 | 301 | 318 | 330 | 0.164 | -0.182 | 0.095 | 0.151 | 0.080 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| GRIP1 |   | 3 | 557 | 696 | 695 | 0.043 | 0.104 | 0.000 | undef | undef |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| NDUFS8 |   | 3 | 557 | 696 | 695 | 0.013 | -0.088 | -0.150 | 0.176 | 0.030 |
|---|
| UBB |   | 3 | 557 | 696 | 695 | 0.146 | -0.027 | 0.012 | undef | -0.019 |
|---|
| ARRDC2 |   | 4 | 440 | 696 | 689 | 0.192 | -0.100 | undef | undef | undef |
|---|
| GRIN2A |   | 12 | 140 | 179 | 192 | -0.113 | -0.068 | 0.162 | 0.125 | undef |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| ZMYND11 |   | 2 | 743 | 696 | 717 | 0.014 | -0.068 | 0.120 | 0.175 | -0.069 |
|---|
| PLAUR |   | 5 | 360 | 696 | 680 | 0.232 | 0.083 | 0.022 | -0.030 | 0.108 |
|---|
| SEPT8 |   | 1 | 1195 | 696 | 744 | 0.209 | 0.148 | 0.097 | 0.077 | 0.169 |
|---|
| C18orf45 |   | 1 | 1195 | 696 | 744 | 0.155 | 0.106 | undef | undef | undef |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| SENP2 |   | 2 | 743 | 696 | 717 | -0.094 | -0.039 | 0.138 | undef | -0.083 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| SNCAIP |   | 10 | 167 | 366 | 362 | -0.091 | 0.013 | 0.092 | 0.136 | 0.059 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6598 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| S phase | GO:0051320 |  | 1.836E-10 | 4.486E-07 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 1.091E-08 | 1.332E-05 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.297E-08 | 1.056E-05 |
|---|
| pigmentation | GO:0043473 |  | 5.06E-08 | 3.09E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.318E-08 | 3.576E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 3.366E-07 | 1.37E-04 |
|---|
| mitochondrial electron transport. NADH to ubiquinone | GO:0006120 |  | 4.996E-07 | 1.744E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 4.999E-07 | 1.527E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.569E-06 | 4.258E-04 |
|---|
| ATP synthesis coupled electron transport | GO:0042773 |  | 1.569E-06 | 3.832E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 2.81E-06 | 6.242E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| S phase | GO:0051320 |  | 2.507E-10 | 6.033E-07 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 1.573E-08 | 1.892E-05 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.87E-08 | 1.5E-05 |
|---|
| pigmentation | GO:0043473 |  | 7.282E-08 | 4.38E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.959E-08 | 4.792E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 4.318E-07 | 1.732E-04 |
|---|
| mitochondrial electron transport. NADH to ubiquinone | GO:0006120 |  | 6.782E-07 | 2.331E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 7.161E-07 | 2.154E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.125E-06 | 5.682E-04 |
|---|
| ATP synthesis coupled electron transport | GO:0042773 |  | 2.125E-06 | 5.114E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 3.394E-06 | 7.424E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| S phase | GO:0051320 |  | 7.51E-08 | 1.727E-04 |
|---|
| pigmentation | GO:0043473 |  | 8.07E-08 | 9.28E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 7.5E-07 | 5.75E-04 |
|---|
| mitochondrial electron transport. NADH to ubiquinone | GO:0006120 |  | 9.226E-07 | 5.305E-04 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 1.088E-06 | 5.007E-04 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.277E-06 | 4.894E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.427E-06 | 1.126E-03 |
|---|
| ATP synthesis coupled electron transport | GO:0042773 |  | 3.427E-06 | 9.853E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.057E-06 | 1.548E-03 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 6.057E-06 | 1.393E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 6.636E-06 | 1.388E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| coagulation | GO:0050817 |  | 1.923E-06 | 3.545E-03 |
|---|
| pigmentation | GO:0043473 |  | 2.443E-06 | 2.251E-03 |
|---|
| hemostasis | GO:0007599 |  | 2.899E-06 | 1.781E-03 |
|---|
| wound healing | GO:0042060 |  | 3.031E-06 | 1.396E-03 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 3.945E-06 | 1.454E-03 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 4.91E-06 | 1.508E-03 |
|---|
| platelet activation | GO:0030168 |  | 6.764E-06 | 1.781E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 9.191E-06 | 2.117E-03 |
|---|
| regulation of body fluid levels | GO:0050878 |  | 1.236E-05 | 2.532E-03 |
|---|
| pigmentation during development | GO:0048066 |  | 1.61E-05 | 2.967E-03 |
|---|
| negative regulation of transcription from RNA polymerase II promoter | GO:0000122 |  | 2.416E-05 | 4.048E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| S phase | GO:0051320 |  | 7.51E-08 | 1.727E-04 |
|---|
| pigmentation | GO:0043473 |  | 8.07E-08 | 9.28E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 7.5E-07 | 5.75E-04 |
|---|
| mitochondrial electron transport. NADH to ubiquinone | GO:0006120 |  | 9.226E-07 | 5.305E-04 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 1.088E-06 | 5.007E-04 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.277E-06 | 4.894E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.427E-06 | 1.126E-03 |
|---|
| ATP synthesis coupled electron transport | GO:0042773 |  | 3.427E-06 | 9.853E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.057E-06 | 1.548E-03 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 6.057E-06 | 1.393E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 6.636E-06 | 1.388E-03 |
|---|



