Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6595
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7538 | 3.519e-03 | 5.730e-04 | 7.708e-01 | 1.554e-06 |
|---|
| Loi | 0.2307 | 7.966e-02 | 1.120e-02 | 4.267e-01 | 3.808e-04 |
|---|
| Schmidt | 0.6735 | 0.000e+00 | 0.000e+00 | 4.480e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.466e-03 | 0.000e+00 |
|---|
| Wang | 0.2620 | 2.618e-03 | 4.327e-02 | 3.938e-01 | 4.461e-05 |
|---|
Expression data for subnetwork 6595 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
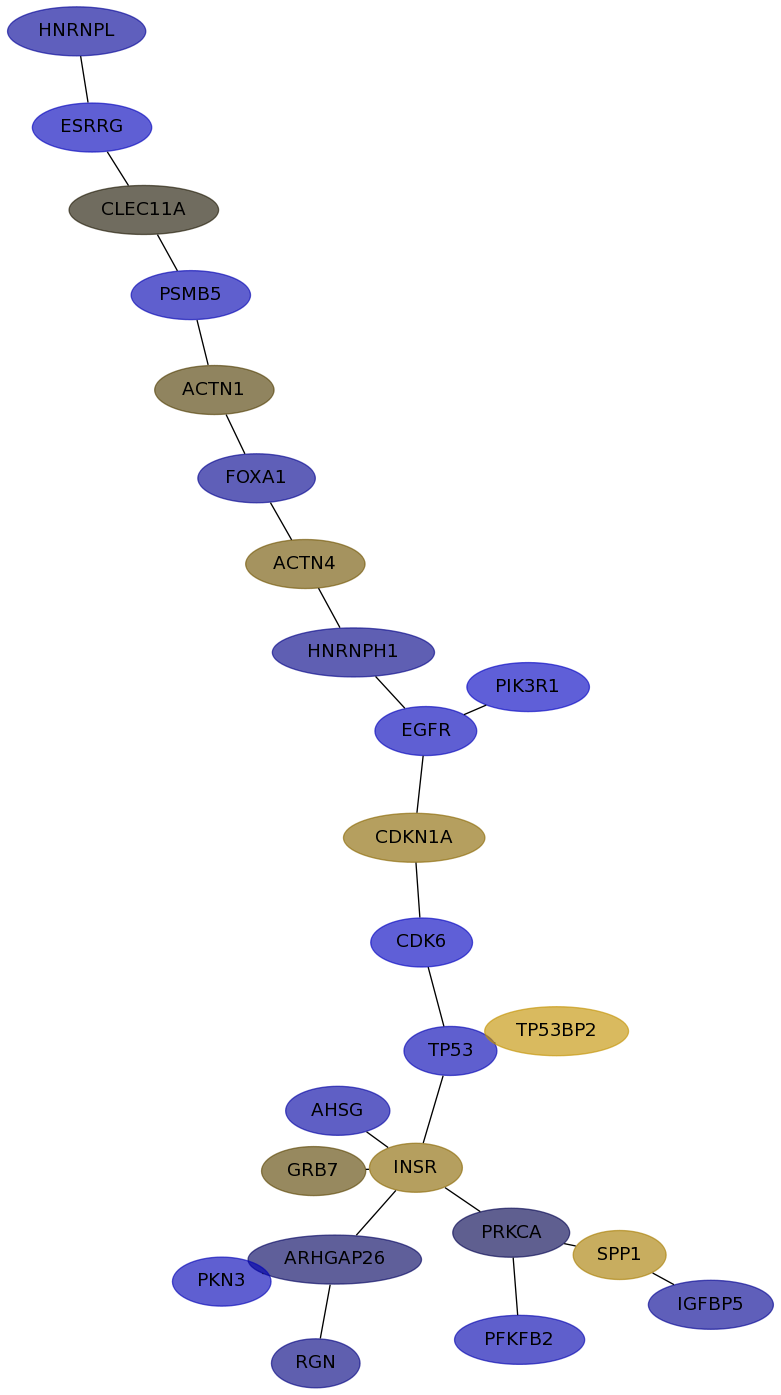
Score for each gene in subnetwork 6595 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| CLEC11A |   | 7 | 256 | 692 | 673 | 0.004 | 0.353 | 0.005 | 0.081 | 0.272 |
|---|
| PSMB5 |   | 2 | 743 | 692 | 713 | -0.163 | 0.195 | undef | 0.137 | undef |
|---|
| PFKFB2 |   | 3 | 557 | 692 | 691 | -0.149 | -0.081 | -0.158 | 0.121 | -0.202 |
|---|
| ESRRG |   | 6 | 301 | 614 | 602 | -0.205 | 0.197 | -0.007 | 0.155 | -0.085 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| HNRNPH1 |   | 9 | 196 | 614 | 597 | -0.059 | 0.248 | 0.125 | 0.144 | -0.133 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| RGN |   | 6 | 301 | 570 | 557 | -0.051 | -0.050 | 0.184 | 0.139 | -0.120 |
|---|
| AHSG |   | 5 | 360 | 412 | 416 | -0.113 | -0.054 | 0.000 | 0.132 | undef |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| HNRNPL |   | 2 | 743 | 614 | 632 | -0.086 | -0.018 | 0.046 | 0.178 | 0.118 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6595 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 7.088E-10 | 1.732E-06 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 4.232E-09 | 5.169E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.774E-08 | 3.887E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.093E-07 | 1.278E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.935E-07 | 1.434E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.006E-07 | 1.631E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 8.134E-07 | 2.839E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 1.593E-06 | 4.864E-04 |
|---|
| negative regulation of glucose import | GO:0046325 |  | 1.593E-06 | 4.324E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.12E-06 | 5.18E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 3.497E-06 | 7.767E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.045E-09 | 2.514E-06 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 6.236E-09 | 7.502E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.689E-08 | 6.167E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.076E-07 | 1.85E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.313E-07 | 2.075E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 5.054E-07 | 2.027E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.089E-06 | 3.744E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 2.132E-06 | 6.412E-04 |
|---|
| negative regulation of glucose import | GO:0046325 |  | 2.132E-06 | 5.7E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.838E-06 | 6.828E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 4.679E-06 | 1.024E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.595E-09 | 3.669E-06 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 5.72E-09 | 6.578E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.919E-08 | 5.305E-05 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.237E-07 | 1.861E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.911E-07 | 1.799E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.565E-07 | 2.133E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.01E-06 | 3.32E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.96E-06 | 8.51E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 2.96E-06 | 7.564E-04 |
|---|
| negative regulation of glucose import | GO:0046325 |  | 2.96E-06 | 6.808E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 5.111E-06 | 1.069E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell motion | GO:0051270 |  | 1.339E-06 | 2.468E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.449E-06 | 2.257E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.549E-06 | 1.566E-03 |
|---|
| positive regulation of endocytosis | GO:0045807 |  | 3.261E-06 | 1.502E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.232E-06 | 1.56E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 4.232E-06 | 1.3E-03 |
|---|
| ossification | GO:0001503 |  | 6.459E-06 | 1.7E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 9.996E-06 | 2.303E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.198E-05 | 2.452E-03 |
|---|
| regulation of cell migration | GO:0030334 |  | 2.857E-05 | 5.266E-03 |
|---|
| glucose homeostasis | GO:0042593 |  | 2.941E-05 | 4.928E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.595E-09 | 3.669E-06 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 5.72E-09 | 6.578E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.919E-08 | 5.305E-05 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.237E-07 | 1.861E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.911E-07 | 1.799E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.565E-07 | 2.133E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.01E-06 | 3.32E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.96E-06 | 8.51E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 2.96E-06 | 7.564E-04 |
|---|
| negative regulation of glucose import | GO:0046325 |  | 2.96E-06 | 6.808E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 5.111E-06 | 1.069E-03 |
|---|



