Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6592
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7540 | 3.507e-03 | 5.700e-04 | 7.704e-01 | 1.540e-06 |
|---|
| Loi | 0.2306 | 7.976e-02 | 1.123e-02 | 4.270e-01 | 3.824e-04 |
|---|
| Schmidt | 0.6736 | 0.000e+00 | 0.000e+00 | 4.476e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.470e-03 | 0.000e+00 |
|---|
| Wang | 0.2620 | 2.620e-03 | 4.329e-02 | 3.939e-01 | 4.467e-05 |
|---|
Expression data for subnetwork 6592 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
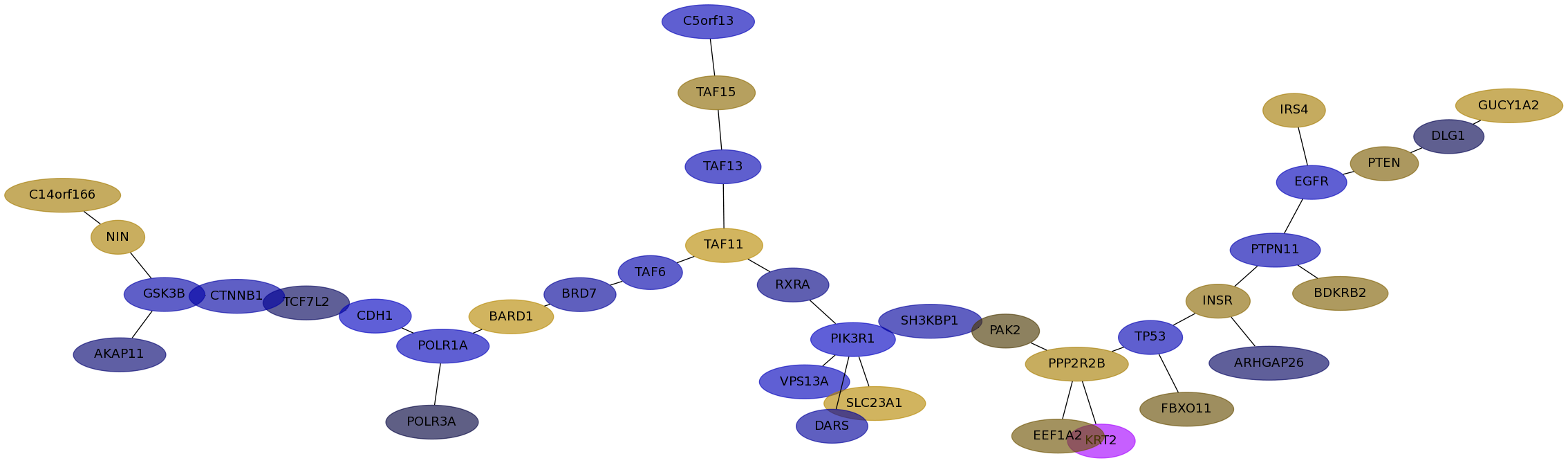
Score for each gene in subnetwork 6592 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| TAF13 |   | 1 | 1195 | 457 | 509 | -0.163 | -0.002 | -0.099 | 0.286 | 0.012 |
|---|
| POLR1A |   | 6 | 301 | 457 | 452 | -0.193 | -0.066 | undef | 0.093 | undef |
|---|
| RXRA |   | 7 | 256 | 457 | 451 | -0.055 | 0.134 | -0.027 | 0.022 | -0.096 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| C5orf13 |   | 1 | 1195 | 457 | 509 | -0.181 | 0.082 | 0.050 | 0.225 | 0.043 |
|---|
| VPS13A |   | 10 | 167 | 236 | 234 | -0.203 | 0.105 | -0.046 | 0.120 | 0.062 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| SH3KBP1 |   | 2 | 743 | 457 | 481 | -0.101 | 0.010 | undef | undef | undef |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| EEF1A2 |   | 8 | 222 | 457 | 450 | 0.032 | 0.220 | -0.119 | 0.018 | 0.102 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| DLG1 |   | 6 | 301 | 457 | 452 | -0.016 | 0.173 | 0.244 | 0.017 | 0.123 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| TAF11 |   | 3 | 557 | 457 | 463 | 0.192 | 0.112 | 0.237 | 0.053 | 0.180 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| AKAP11 |   | 4 | 440 | 457 | 458 | -0.034 | -0.099 | -0.038 | 0.251 | 0.017 |
|---|
| SLC23A1 |   | 2 | 743 | 457 | 481 | 0.179 | 0.104 | undef | 0.052 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| C14orf166 |   | 1 | 1195 | 457 | 509 | 0.116 | 0.108 | 0.106 | 0.078 | -0.126 |
|---|
| GUCY1A2 |   | 2 | 743 | 457 | 481 | 0.135 | -0.058 | -0.088 | 0.307 | 0.131 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| POLR3A |   | 8 | 222 | 296 | 287 | -0.010 | -0.032 | undef | 0.091 | undef |
|---|
| NIN |   | 1 | 1195 | 457 | 509 | 0.135 | -0.034 | -0.088 | 0.152 | 0.082 |
|---|
| FBXO11 |   | 5 | 360 | 457 | 455 | 0.027 | 0.186 | -0.069 | 0.238 | 0.149 |
|---|
| TAF15 |   | 2 | 743 | 457 | 481 | 0.066 | 0.089 | 0.207 | 0.117 | 0.095 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| DARS |   | 1 | 1195 | 457 | 509 | -0.097 | -0.007 | -0.156 | undef | 0.181 |
|---|
| KRT2 |   | 2 | 743 | 457 | 481 | undef | -0.004 | 0.229 | -0.004 | -0.094 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| TAF6 |   | 2 | 743 | 457 | 481 | -0.139 | -0.013 | -0.015 | 0.203 | 0.073 |
|---|
| BDKRB2 |   | 1 | 1195 | 457 | 509 | 0.049 | -0.025 | 0.128 | 0.197 | -0.054 |
|---|
GO Enrichment output for subnetwork 6592 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.799E-15 | 1.905E-11 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.093E-13 | 6.221E-10 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 2.927E-11 | 2.384E-08 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 4.921E-11 | 3.006E-08 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 8.562E-11 | 4.183E-08 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.68E-10 | 6.842E-08 |
|---|
| regulation of insulin secretion | GO:0050796 |  | 1.88E-10 | 6.562E-08 |
|---|
| regulation of peptide secretion | GO:0002791 |  | 4.99E-10 | 1.524E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 5.561E-10 | 1.509E-07 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 8.056E-10 | 1.968E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.664E-09 | 3.695E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.149E-14 | 2.766E-11 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.419E-13 | 4.113E-10 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 4.306E-11 | 3.453E-08 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 6.312E-11 | 3.797E-08 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.334E-10 | 6.419E-08 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.616E-10 | 1.049E-07 |
|---|
| regulation of insulin secretion | GO:0050796 |  | 2.763E-10 | 9.496E-08 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 7.228E-10 | 2.174E-07 |
|---|
| regulation of peptide secretion | GO:0002791 |  | 7.326E-10 | 1.959E-07 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.252E-09 | 3.012E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.162E-09 | 4.73E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.552E-15 | 3.569E-12 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.095E-11 | 3.56E-08 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.218E-10 | 9.339E-08 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 3.804E-10 | 2.188E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 7.15E-10 | 3.289E-07 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 1.596E-09 | 6.117E-07 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 2.85E-09 | 9.364E-07 |
|---|
| organ growth | GO:0035265 |  | 4.979E-09 | 1.431E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 4.979E-09 | 1.272E-06 |
|---|
| regulation of protein transport | GO:0051223 |  | 8.987E-09 | 2.067E-06 |
|---|
| regulation of insulin secretion | GO:0050796 |  | 9.27E-09 | 1.938E-06 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.85E-08 | 7.095E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 5.817E-08 | 5.36E-05 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.211E-07 | 7.437E-05 |
|---|
| ER overload response | GO:0006983 |  | 1.198E-06 | 5.519E-04 |
|---|
| regulation of protein transport | GO:0051223 |  | 1.923E-06 | 7.087E-04 |
|---|
| regulation of protein localization | GO:0032880 |  | 2.774E-06 | 8.521E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 4.203E-06 | 1.107E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 9.748E-06 | 2.246E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 9.748E-06 | 1.996E-03 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 9.748E-06 | 1.797E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.182E-05 | 1.981E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.552E-15 | 3.569E-12 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.095E-11 | 3.56E-08 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.218E-10 | 9.339E-08 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 3.804E-10 | 2.188E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 7.15E-10 | 3.289E-07 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 1.596E-09 | 6.117E-07 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 2.85E-09 | 9.364E-07 |
|---|
| organ growth | GO:0035265 |  | 4.979E-09 | 1.431E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 4.979E-09 | 1.272E-06 |
|---|
| regulation of protein transport | GO:0051223 |  | 8.987E-09 | 2.067E-06 |
|---|
| regulation of insulin secretion | GO:0050796 |  | 9.27E-09 | 1.938E-06 |
|---|



