Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6571
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7543 | 3.488e-03 | 5.660e-04 | 7.696e-01 | 1.519e-06 |
|---|
| Loi | 0.2306 | 7.984e-02 | 1.125e-02 | 4.272e-01 | 3.836e-04 |
|---|
| Schmidt | 0.6733 | 0.000e+00 | 0.000e+00 | 4.508e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.470e-03 | 0.000e+00 |
|---|
| Wang | 0.2619 | 2.641e-03 | 4.347e-02 | 3.948e-01 | 4.533e-05 |
|---|
Expression data for subnetwork 6571 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
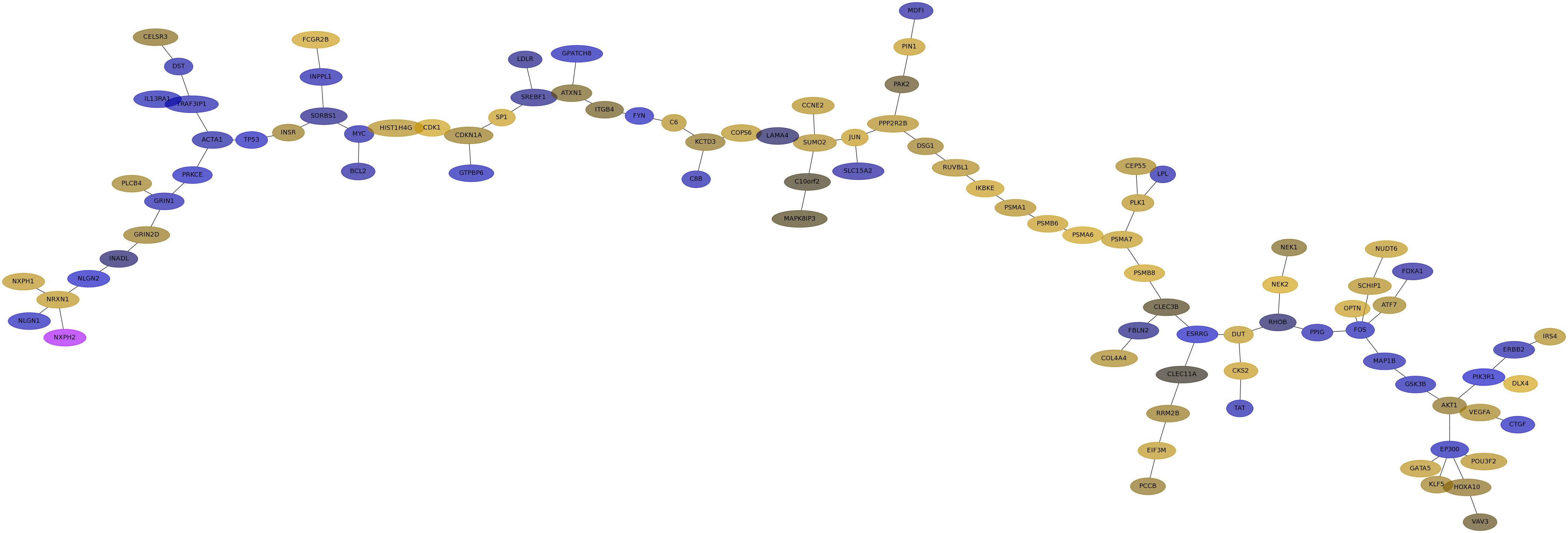
Score for each gene in subnetwork 6571 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| CCNE2 |   | 11 | 148 | 412 | 407 | 0.136 | -0.178 | 0.241 | undef | -0.055 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| FBLN2 |   | 5 | 360 | 83 | 110 | -0.036 | 0.122 | 0.207 | 0.037 | -0.007 |
|---|
| CELSR3 |   | 1 | 1195 | 928 | 965 | 0.040 | -0.022 | 0.055 | undef | -0.231 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| LDLR |   | 5 | 360 | 780 | 767 | -0.041 | 0.218 | 0.039 | -0.028 | 0.064 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| IL13RA1 |   | 1 | 1195 | 928 | 965 | -0.131 | 0.311 | -0.016 | 0.252 | 0.069 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| NRXN1 |   | 2 | 743 | 928 | 929 | 0.171 | -0.004 | -0.097 | undef | -0.033 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| PSMA6 |   | 5 | 360 | 842 | 828 | 0.280 | -0.118 | -0.018 | undef | -0.091 |
|---|
| PSMB8 |   | 3 | 557 | 928 | 925 | 0.254 | -0.071 | -0.088 | undef | -0.139 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| PSMA7 |   | 2 | 743 | 928 | 929 | 0.188 | -0.067 | 0.223 | -0.022 | -0.001 |
|---|
| DUT |   | 1 | 1195 | 928 | 965 | 0.177 | -0.047 | 0.346 | undef | 0.080 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| CKS2 |   | 9 | 196 | 928 | 903 | 0.214 | -0.220 | 0.089 | -0.003 | -0.056 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| TAT |   | 11 | 148 | 83 | 97 | -0.113 | 0.030 | -0.182 | 0.037 | -0.074 |
|---|
| PLK1 |   | 11 | 148 | 236 | 232 | 0.142 | -0.143 | 0.094 | -0.192 | -0.163 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| OPTN |   | 7 | 256 | 1 | 47 | 0.226 | -0.099 | 0.015 | undef | 0.035 |
|---|
| INADL |   | 2 | 743 | 928 | 929 | -0.019 | -0.045 | 0.061 | -0.011 | 0.027 |
|---|
| FYN |   | 10 | 167 | 1 | 40 | -0.171 | -0.167 | 0.073 | -0.137 | 0.110 |
|---|
| GTPBP6 |   | 6 | 301 | 928 | 907 | -0.143 | -0.131 | -0.167 | 0.155 | -0.129 |
|---|
| RUVBL1 |   | 2 | 743 | 658 | 674 | 0.112 | -0.107 | 0.049 | -0.179 | 0.027 |
|---|
| NXPH1 |   | 2 | 743 | 928 | 929 | 0.162 | 0.109 | undef | undef | undef |
|---|
| CEP55 |   | 2 | 743 | 928 | 929 | 0.097 | -0.196 | 0.056 | -0.002 | 0.052 |
|---|
| TRAF3IP1 |   | 9 | 196 | 83 | 99 | -0.113 | -0.063 | -0.088 | 0.276 | undef |
|---|
| ACTA1 |   | 4 | 440 | 83 | 112 | -0.090 | -0.041 | 0.145 | 0.248 | 0.132 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| DSG1 |   | 10 | 167 | 658 | 636 | 0.072 | -0.075 | 0.252 | -0.061 | 0.068 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| GATA5 |   | 8 | 222 | 366 | 373 | 0.161 | -0.031 | undef | undef | undef |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| NUDT6 |   | 1 | 1195 | 928 | 965 | 0.181 | -0.165 | 0.195 | undef | 0.149 |
|---|
| INPPL1 |   | 6 | 301 | 141 | 152 | -0.122 | 0.072 | -0.087 | 0.174 | 0.156 |
|---|
| C6 |   | 4 | 440 | 913 | 904 | 0.128 | 0.055 | 0.169 | 0.161 | 0.054 |
|---|
| C10orf2 |   | 2 | 743 | 366 | 395 | 0.007 | 0.040 | 0.228 | undef | -0.028 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| MAPK8IP3 |   | 3 | 557 | 928 | 925 | 0.010 | 0.045 | -0.046 | undef | 0.059 |
|---|
| C8B |   | 1 | 1195 | 928 | 965 | -0.111 | 0.135 | -0.089 | -0.050 | -0.092 |
|---|
| HOXA10 |   | 4 | 440 | 1 | 59 | 0.046 | 0.203 | -0.057 | -0.160 | 0.016 |
|---|
| PRKCE |   | 2 | 743 | 928 | 929 | -0.166 | 0.241 | -0.088 | 0.142 | -0.101 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| NLGN2 |   | 2 | 743 | 928 | 929 | -0.217 | 0.241 | undef | undef | undef |
|---|
| PPIG |   | 8 | 222 | 648 | 631 | -0.116 | 0.024 | 0.221 | -0.065 | 0.101 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| SCHIP1 |   | 10 | 167 | 478 | 459 | 0.130 | 0.136 | 0.105 | -0.022 | 0.198 |
|---|
| GPATCH8 |   | 8 | 222 | 614 | 599 | -0.143 | 0.014 | 0.019 | -0.070 | 0.061 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| NEK2 |   | 8 | 222 | 141 | 149 | 0.292 | -0.106 | 0.020 | 0.083 | 0.165 |
|---|
| SREBF1 |   | 9 | 196 | 478 | 460 | -0.042 | 0.232 | -0.195 | 0.140 | -0.074 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| DLX4 |   | 10 | 167 | 236 | 234 | 0.291 | 0.057 | -0.004 | undef | -0.059 |
|---|
| KCTD3 |   | 3 | 557 | 913 | 908 | 0.051 | 0.016 | 0.086 | 0.256 | 0.053 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| PSMB6 |   | 1 | 1195 | 928 | 965 | 0.207 | -0.046 | 0.106 | undef | -0.003 |
|---|
| FCGR2B |   | 2 | 743 | 913 | 914 | 0.253 | -0.024 | 0.051 | -0.284 | -0.002 |
|---|
| VAV3 |   | 9 | 196 | 638 | 622 | 0.015 | 0.071 | -0.245 | 0.148 | 0.038 |
|---|
| PCCB |   | 1 | 1195 | 928 | 965 | 0.050 | -0.026 | -0.089 | -0.080 | -0.003 |
|---|
| CLEC11A |   | 7 | 256 | 692 | 673 | 0.004 | 0.353 | 0.005 | 0.081 | 0.272 |
|---|
| NXPH2 |   | 1 | 1195 | 928 | 965 | undef | 0.164 | undef | 0.039 | undef |
|---|
| COL4A4 |   | 6 | 301 | 83 | 105 | 0.102 | 0.155 | -0.039 | 0.295 | 0.068 |
|---|
| ESRRG |   | 6 | 301 | 614 | 602 | -0.205 | 0.197 | -0.007 | 0.155 | -0.085 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| GRIN1 |   | 3 | 557 | 928 | 925 | -0.114 | -0.067 | -0.111 | undef | 0.115 |
|---|
| PSMA1 |   | 2 | 743 | 806 | 806 | 0.129 | -0.118 | 0.125 | undef | -0.082 |
|---|
| NLGN1 |   | 2 | 743 | 928 | 929 | -0.163 | 0.025 | 0.150 | 0.263 | -0.066 |
|---|
| HIST1H4G |   | 1 | 1195 | 928 | 965 | 0.135 | 0.043 | -0.070 | 0.073 | 0.030 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| RHOB |   | 1 | 1195 | 928 | 965 | -0.018 | 0.109 | -0.008 | 0.043 | 0.015 |
|---|
| SLC15A2 |   | 4 | 440 | 928 | 912 | -0.076 | 0.109 | -0.068 | 0.050 | -0.123 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| IKBKE |   | 10 | 167 | 552 | 539 | 0.227 | -0.041 | -0.095 | -0.197 | 0.079 |
|---|
| RRM2B |   | 1 | 1195 | 928 | 965 | 0.056 | -0.107 | undef | undef | undef |
|---|
| EIF3M |   | 1 | 1195 | 928 | 965 | 0.183 | 0.028 | 0.021 | -0.102 | 0.084 |
|---|
| PLCB4 |   | 4 | 440 | 928 | 912 | 0.074 | 0.179 | -0.113 | 0.248 | -0.020 |
|---|
| GRIN2D |   | 13 | 124 | 552 | 522 | 0.058 | 0.155 | 0.000 | undef | undef |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| CLEC3B |   | 2 | 743 | 928 | 929 | 0.009 | -0.021 | 0.139 | undef | 0.033 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6571 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 1.078E-08 | 2.633E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.562E-08 | 3.13E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 3.604E-08 | 2.934E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 3.604E-08 | 2.201E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 6.151E-08 | 3.005E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 7.537E-08 | 3.069E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.223E-07 | 4.268E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.085E-07 | 6.368E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.474E-07 | 1.214E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.044E-06 | 2.551E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.055E-06 | 4.563E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 1.508E-08 | 3.628E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 4.842E-08 | 5.825E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 6.799E-08 | 5.453E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 6.799E-08 | 4.089E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.158E-07 | 5.57E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.417E-07 | 5.682E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 2.293E-07 | 7.882E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.15E-07 | 9.473E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 7.789E-07 | 2.082E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.288E-06 | 3.1E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.863E-06 | 6.262E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.678E-07 | 3.859E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.442E-07 | 2.808E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.442E-07 | 1.872E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.912E-07 | 2.249E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 4.895E-07 | 2.251E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.914E-07 | 2.65E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 7.505E-07 | 2.466E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.29E-06 | 6.583E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.269E-06 | 8.353E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 8.581E-06 | 1.974E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 9.387E-06 | 1.963E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| T cell lineage commitment | GO:0002360 |  | 5.619E-06 | 0.01035656 |
|---|
| cell junction assembly | GO:0034329 |  | 9.975E-06 | 9.192E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.117E-05 | 6.863E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 1.117E-05 | 5.147E-03 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 1.117E-05 | 4.118E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 1.117E-05 | 3.431E-03 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 1.45E-05 | 3.817E-03 |
|---|
| response to gamma radiation | GO:0010332 |  | 1.943E-05 | 4.477E-03 |
|---|
| regulation of mitotic cell cycle | GO:0007346 |  | 2.192E-05 | 4.488E-03 |
|---|
| cell-substrate adhesion | GO:0031589 |  | 2.395E-05 | 4.413E-03 |
|---|
| cell junction organization | GO:0034330 |  | 2.971E-05 | 4.978E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.678E-07 | 3.859E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.442E-07 | 2.808E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.442E-07 | 1.872E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.912E-07 | 2.249E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 4.895E-07 | 2.251E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.914E-07 | 2.65E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 7.505E-07 | 2.466E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.29E-06 | 6.583E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.269E-06 | 8.353E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 8.581E-06 | 1.974E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 9.387E-06 | 1.963E-03 |
|---|



