Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6565
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7541 | 3.499e-03 | 5.690e-04 | 7.700e-01 | 1.533e-06 |
|---|
| Loi | 0.2306 | 7.977e-02 | 1.123e-02 | 4.270e-01 | 3.824e-04 |
|---|
| Schmidt | 0.6731 | 0.000e+00 | 0.000e+00 | 4.525e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.458e-03 | 0.000e+00 |
|---|
| Wang | 0.2618 | 2.647e-03 | 4.353e-02 | 3.950e-01 | 4.551e-05 |
|---|
Expression data for subnetwork 6565 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
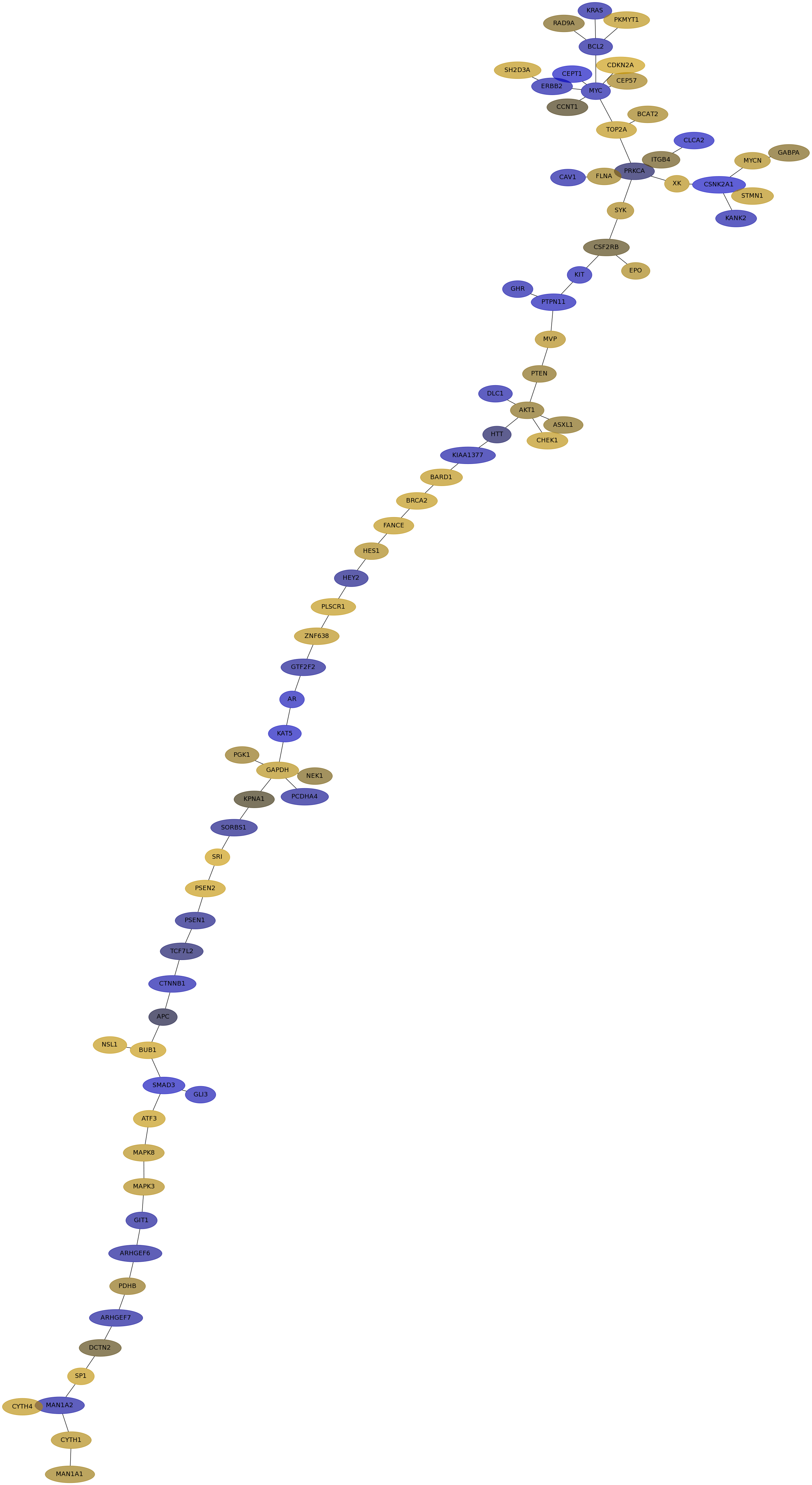
Subnetwork structure for each dataset
- IPC
Score for each gene in subnetwork 6565 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| MAPK8 |   | 8 | 222 | 696 | 677 | 0.153 | 0.200 | 0.249 | -0.066 | 0.138 |
|---|
| RAD9A |   | 2 | 743 | 1510 | 1502 | 0.033 | -0.044 | 0.014 | 0.279 | -0.051 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| DLC1 |   | 1 | 1195 | 1510 | 1530 | -0.101 | 0.088 | 0.245 | 0.069 | 0.258 |
|---|
| KAT5 |   | 1 | 1195 | 1510 | 1530 | -0.190 | -0.060 | -0.026 | -0.089 | 0.085 |
|---|
| CSNK2A1 |   | 3 | 557 | 1510 | 1486 | -0.215 | 0.029 | undef | 0.122 | undef |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| APC |   | 1 | 1195 | 1510 | 1530 | -0.006 | -0.004 | 0.067 | -0.002 | 0.139 |
|---|
| GIT1 |   | 2 | 743 | 1510 | 1502 | -0.067 | -0.099 | -0.045 | undef | -0.065 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| STMN1 |   | 4 | 440 | 1461 | 1437 | 0.168 | -0.033 | 0.198 | 0.105 | 0.062 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| BCAT2 |   | 3 | 557 | 1510 | 1486 | 0.091 | -0.103 | -0.096 | -0.128 | -0.030 |
|---|
| HEY2 |   | 8 | 222 | 570 | 555 | -0.045 | -0.021 | 0.398 | -0.024 | -0.058 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| SRI |   | 7 | 256 | 614 | 601 | 0.270 | -0.040 | 0.181 | 0.185 | 0.057 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| ZNF638 |   | 5 | 360 | 806 | 784 | 0.164 | 0.088 | 0.020 | 0.077 | -0.045 |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| ASXL1 |   | 2 | 743 | 1510 | 1502 | 0.044 | 0.146 | 0.031 | undef | 0.041 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| PKMYT1 |   | 7 | 256 | 1510 | 1463 | 0.175 | -0.143 | 0.046 | 0.105 | -0.070 |
|---|
| CAV1 |   | 2 | 743 | 1510 | 1502 | -0.092 | 0.124 | 0.079 | undef | 0.119 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| FANCE |   | 1 | 1195 | 1510 | 1530 | 0.184 | -0.209 | 0.079 | undef | 0.106 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| SMAD3 |   | 3 | 557 | 842 | 834 | -0.176 | 0.063 | 0.239 | -0.068 | -0.012 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| NSL1 |   | 2 | 743 | 780 | 787 | 0.221 | -0.079 | 0.288 | undef | 0.174 |
|---|
| ARHGEF6 |   | 10 | 167 | 525 | 507 | -0.068 | -0.120 | 0.166 | undef | -0.115 |
|---|
| CCNT1 |   | 6 | 301 | 478 | 473 | 0.008 | -0.028 | 0.162 | 0.007 | -0.213 |
|---|
| CLCA2 |   | 9 | 196 | 412 | 411 | -0.176 | 0.243 | -0.153 | 0.191 | -0.000 |
|---|
| CYTH4 |   | 1 | 1195 | 1510 | 1530 | 0.185 | -0.105 | 0.025 | -0.154 | -0.052 |
|---|
| PCDHA4 |   | 2 | 743 | 1510 | 1502 | -0.060 | 0.110 | -0.192 | 0.290 | -0.029 |
|---|
| SH2D3A |   | 3 | 557 | 1510 | 1486 | 0.197 | 0.141 | -0.106 | -0.012 | 0.121 |
|---|
| MAN1A2 |   | 1 | 1195 | 1510 | 1530 | -0.087 | 0.152 | 0.133 | 0.118 | 0.070 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| DCTN2 |   | 8 | 222 | 366 | 373 | 0.014 | 0.150 | 0.061 | 0.113 | 0.303 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| PSEN1 |   | 10 | 167 | 366 | 362 | -0.041 | 0.110 | undef | 0.083 | undef |
|---|
| MAN1A1 |   | 1 | 1195 | 1510 | 1530 | 0.085 | 0.007 | 0.058 | undef | 0.069 |
|---|
| CEP57 |   | 1 | 1195 | 1510 | 1530 | 0.091 | 0.097 | 0.219 | 0.094 | 0.027 |
|---|
| CSF2RB |   | 3 | 557 | 1492 | 1464 | 0.013 | -0.196 | -0.109 | 0.012 | -0.141 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| GTF2F2 |   | 4 | 440 | 957 | 942 | -0.059 | -0.085 | 0.119 | -0.004 | -0.054 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| BUB1 |   | 6 | 301 | 590 | 582 | 0.246 | -0.172 | 0.013 | -0.007 | 0.170 |
|---|
| KANK2 |   | 1 | 1195 | 1510 | 1530 | -0.104 | 0.122 | 0.099 | 0.094 | 0.218 |
|---|
| MYCN |   | 7 | 256 | 764 | 756 | 0.125 | 0.057 | -0.003 | -0.081 | -0.084 |
|---|
| MVP |   | 1 | 1195 | 1510 | 1530 | 0.137 | -0.213 | -0.132 | -0.003 | 0.096 |
|---|
| SYK |   | 2 | 743 | 1510 | 1502 | 0.106 | -0.251 | -0.134 | 0.017 | 0.163 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| CYTH1 |   | 1 | 1195 | 1510 | 1530 | 0.142 | -0.319 | 0.134 | 0.024 | 0.105 |
|---|
| HES1 |   | 4 | 440 | 570 | 565 | 0.115 | 0.157 | 0.010 | 0.214 | -0.106 |
|---|
| PLSCR1 |   | 10 | 167 | 412 | 408 | 0.202 | -0.159 | 0.091 | undef | -0.105 |
|---|
| KRAS |   | 2 | 743 | 1510 | 1502 | -0.083 | -0.127 | -0.030 | undef | 0.182 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| MAPK3 |   | 6 | 301 | 141 | 152 | 0.137 | -0.102 | -0.126 | undef | -0.086 |
|---|
| KPNA1 |   | 9 | 196 | 141 | 147 | 0.007 | 0.152 | -0.083 | 0.073 | 0.086 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| PGK1 |   | 1 | 1195 | 1510 | 1530 | 0.059 | 0.095 | 0.115 | 0.143 | 0.119 |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| CEPT1 |   | 2 | 743 | 1510 | 1502 | -0.208 | 0.038 | 0.194 | 0.163 | 0.205 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| EPO |   | 4 | 440 | 1492 | 1461 | 0.109 | 0.086 | -0.083 | -0.048 | 0.155 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6565 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.032E-13 | 4.964E-10 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.755E-11 | 5.809E-08 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 6.968E-11 | 5.674E-08 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.231E-10 | 1.973E-07 |
|---|
| organ growth | GO:0035265 |  | 3.231E-10 | 1.579E-07 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.673E-10 | 1.903E-07 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.009E-09 | 3.52E-07 |
|---|
| regulation of protein localization | GO:0032880 |  | 1.621E-09 | 4.95E-07 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 2.666E-09 | 7.235E-07 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 4.094E-09 | 1E-06 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 5.98E-09 | 1.328E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.214E-13 | 7.733E-10 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.829E-11 | 3.404E-08 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 7.867E-11 | 6.31E-08 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 4.031E-10 | 2.425E-07 |
|---|
| organ growth | GO:0035265 |  | 4.031E-10 | 1.94E-07 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 6.563E-10 | 2.632E-07 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.415E-09 | 4.864E-07 |
|---|
| regulation of protein localization | GO:0032880 |  | 2.438E-09 | 7.332E-07 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 3.461E-09 | 9.252E-07 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.553E-09 | 8.548E-07 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 6.815E-09 | 1.491E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.161E-10 | 7.27E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.284E-10 | 7.227E-07 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 6.44E-10 | 4.937E-07 |
|---|
| organ growth | GO:0035265 |  | 1.098E-09 | 6.316E-07 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 3.064E-09 | 1.409E-06 |
|---|
| regulation of protein localization | GO:0032880 |  | 4.224E-09 | 1.619E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 5.154E-09 | 1.694E-06 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.11E-08 | 3.192E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 3.03E-08 | 7.742E-06 |
|---|
| glycolysis | GO:0006096 |  | 1.143E-07 | 2.63E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.091E-07 | 4.373E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.199E-08 | 4.053E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 3.634E-08 | 3.348E-05 |
|---|
| focal adhesion formation | GO:0048041 |  | 4.305E-08 | 2.645E-05 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 8.832E-08 | 4.07E-05 |
|---|
| regulation of protein localization | GO:0032880 |  | 1.838E-07 | 6.776E-05 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 3.556E-07 | 1.092E-04 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 5.894E-07 | 1.552E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 9.21E-07 | 2.122E-04 |
|---|
| gland development | GO:0048732 |  | 1.512E-06 | 3.097E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.512E-06 | 2.787E-04 |
|---|
| regulation of protein transport | GO:0051223 |  | 2.427E-06 | 4.066E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.161E-10 | 7.27E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.284E-10 | 7.227E-07 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 6.44E-10 | 4.937E-07 |
|---|
| organ growth | GO:0035265 |  | 1.098E-09 | 6.316E-07 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 3.064E-09 | 1.409E-06 |
|---|
| regulation of protein localization | GO:0032880 |  | 4.224E-09 | 1.619E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 5.154E-09 | 1.694E-06 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.11E-08 | 3.192E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 3.03E-08 | 7.742E-06 |
|---|
| glycolysis | GO:0006096 |  | 1.143E-07 | 2.63E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.091E-07 | 4.373E-05 |
|---|



