Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6542
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7547 | 3.464e-03 | 5.610e-04 | 7.686e-01 | 1.494e-06 |
|---|
| Loi | 0.2305 | 7.991e-02 | 1.127e-02 | 4.274e-01 | 3.847e-04 |
|---|
| Schmidt | 0.6727 | 0.000e+00 | 0.000e+00 | 4.565e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.469e-03 | 0.000e+00 |
|---|
| Wang | 0.2616 | 2.679e-03 | 4.382e-02 | 3.964e-01 | 4.654e-05 |
|---|
Expression data for subnetwork 6542 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
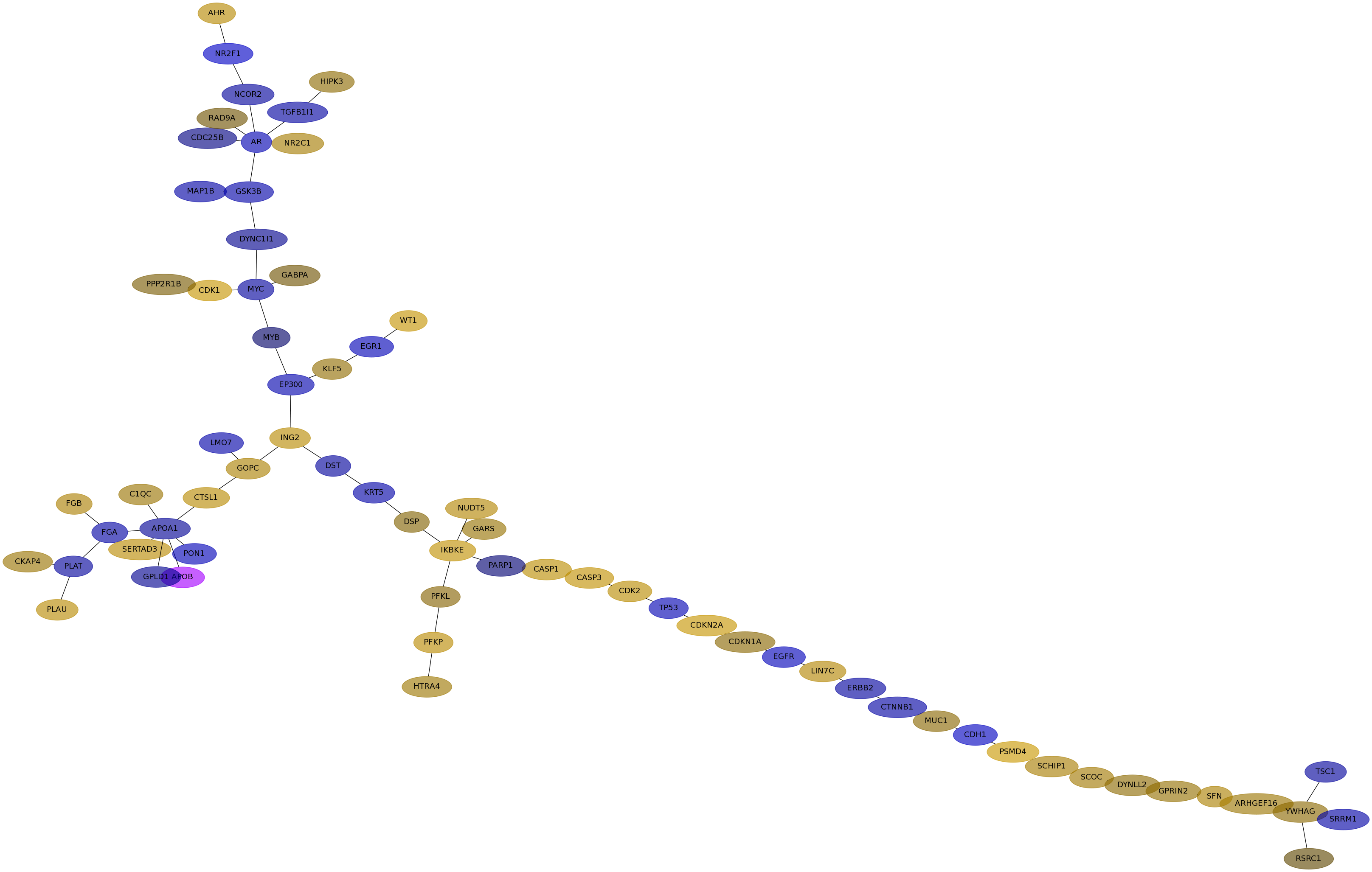
Score for each gene in subnetwork 6542 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| LMO7 |   | 2 | 743 | 1735 | 1721 | -0.129 | 0.068 | 0.000 | 0.197 | 0.151 |
|---|
| CASP3 |   | 11 | 148 | 141 | 143 | 0.234 | -0.032 | -0.142 | -0.097 | -0.084 |
|---|
| RAD9A |   | 2 | 743 | 1510 | 1502 | 0.033 | -0.044 | 0.014 | 0.279 | -0.051 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PLAT |   | 1 | 1195 | 1771 | 1788 | -0.096 | 0.089 | 0.090 | 0.013 | 0.023 |
|---|
| PPP2R1B |   | 6 | 301 | 141 | 152 | 0.042 | 0.002 | -0.002 | undef | 0.112 |
|---|
| ARHGEF16 |   | 10 | 167 | 296 | 283 | 0.096 | 0.036 | -0.047 | 0.200 | 0.038 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| PLAU |   | 12 | 140 | 179 | 192 | 0.187 | 0.153 | 0.141 | 0.072 | 0.192 |
|---|
| C1QC |   | 1 | 1195 | 1771 | 1788 | 0.093 | -0.269 | undef | undef | undef |
|---|
| CDC25B |   | 2 | 743 | 1771 | 1754 | -0.054 | -0.075 | -0.037 | undef | 0.009 |
|---|
| NR2C1 |   | 8 | 222 | 727 | 703 | 0.121 | 0.039 | 0.119 | -0.197 | -0.026 |
|---|
| GARS |   | 2 | 743 | 1222 | 1215 | 0.087 | 0.149 | -0.237 | 0.171 | 0.045 |
|---|
| KRT5 |   | 10 | 167 | 806 | 780 | -0.125 | 0.046 | 0.064 | -0.006 | -0.190 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| GOPC |   | 1 | 1195 | 1771 | 1788 | 0.140 | -0.025 | undef | -0.071 | undef |
|---|
| SERTAD3 |   | 1 | 1195 | 1771 | 1788 | 0.195 | 0.095 | -0.040 | 0.048 | 0.072 |
|---|
| DSP |   | 7 | 256 | 590 | 570 | 0.054 | 0.235 | -0.119 | -0.033 | 0.053 |
|---|
| PON1 |   | 1 | 1195 | 1771 | 1788 | -0.182 | 0.160 | 0.086 | 0.178 | -0.114 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| HTRA4 |   | 8 | 222 | 83 | 101 | 0.102 | undef | undef | undef | undef |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| PFKP |   | 6 | 301 | 83 | 105 | 0.195 | -0.076 | -0.040 | undef | 0.094 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| SCOC |   | 1 | 1195 | 1771 | 1788 | 0.111 | -0.057 | undef | undef | undef |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| SRRM1 |   | 10 | 167 | 296 | 283 | -0.120 | 0.139 | 0.244 | 0.294 | -0.109 |
|---|
| APOB |   | 3 | 557 | 1771 | 1745 | undef | 0.001 | 0.156 | 0.151 | 0.144 |
|---|
| DYNC1I1 |   | 5 | 360 | 614 | 612 | -0.069 | 0.013 | -0.086 | -0.045 | 0.088 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| PFKL |   | 7 | 256 | 83 | 102 | 0.056 | 0.106 | -0.173 | 0.160 | -0.158 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| SCHIP1 |   | 10 | 167 | 478 | 459 | 0.130 | 0.136 | 0.105 | -0.022 | 0.198 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| NUDT5 |   | 1 | 1195 | 1771 | 1788 | 0.165 | -0.298 | undef | 0.138 | undef |
|---|
| CASP1 |   | 6 | 301 | 552 | 544 | 0.204 | -0.166 | -0.046 | undef | -0.034 |
|---|
| CKAP4 |   | 6 | 301 | 780 | 764 | 0.093 | 0.152 | -0.167 | 0.315 | -0.017 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| ING2 |   | 3 | 557 | 1 | 63 | 0.189 | 0.168 | 0.051 | 0.026 | -0.054 |
|---|
| PSMD4 |   | 11 | 148 | 296 | 282 | 0.285 | -0.243 | -0.043 | 0.087 | -0.045 |
|---|
| LIN7C |   | 10 | 167 | 590 | 566 | 0.166 | 0.088 | 0.118 | -0.018 | 0.094 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| GPLD1 |   | 1 | 1195 | 1771 | 1788 | -0.073 | 0.116 | -0.087 | 0.080 | 0.057 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| CTSL1 |   | 1 | 1195 | 1771 | 1788 | 0.187 | -0.080 | 0.091 | -0.095 | 0.178 |
|---|
| TSC1 |   | 2 | 743 | 1564 | 1549 | -0.094 | 0.257 | 0.103 | -0.042 | 0.077 |
|---|
| IKBKE |   | 10 | 167 | 552 | 539 | 0.227 | -0.041 | -0.095 | -0.197 | 0.079 |
|---|
| DYNLL2 |   | 2 | 743 | 1735 | 1721 | 0.068 | -0.127 | undef | undef | undef |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| FGB |   | 1 | 1195 | 1771 | 1788 | 0.136 | 0.231 | -0.035 | undef | -0.016 |
|---|
| FGA |   | 1 | 1195 | 1771 | 1788 | -0.114 | 0.080 | -0.088 | -0.026 | 0.132 |
|---|
| APOA1 |   | 1 | 1195 | 1771 | 1788 | -0.083 | 0.041 | 0.187 | 0.054 | -0.019 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| HIPK3 |   | 9 | 196 | 1461 | 1417 | 0.069 | 0.005 | -0.027 | undef | 0.175 |
|---|
| AHR |   | 8 | 222 | 478 | 461 | 0.180 | 0.067 | 0.091 | -0.182 | -0.129 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6542 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.354E-10 | 8.194E-07 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.926E-09 | 4.795E-06 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.463E-08 | 2.82E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.832E-08 | 2.341E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.664E-08 | 2.768E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.404E-08 | 2.607E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 9.2E-08 | 3.211E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.441E-07 | 4.399E-05 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 2.153E-07 | 5.844E-05 |
|---|
| negative regulation of mast cell proliferation | GO:0070667 |  | 2.153E-07 | 5.26E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.725E-07 | 6.052E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.851E-10 | 1.167E-06 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 5.351E-09 | 6.437E-06 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.271E-08 | 3.425E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.741E-08 | 2.852E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 7.087E-08 | 3.41E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.206E-08 | 3.692E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.181E-07 | 4.058E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.181E-07 | 3.551E-05 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 2.762E-07 | 7.383E-05 |
|---|
| negative regulation of mast cell proliferation | GO:0070667 |  | 2.762E-07 | 6.644E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.906E-07 | 8.544E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.303E-09 | 2.997E-06 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.251E-08 | 2.589E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.262E-07 | 9.676E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.393E-07 | 1.376E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.748E-07 | 1.724E-04 |
|---|
| negative regulation of mast cell proliferation | GO:0070667 |  | 5.859E-07 | 2.246E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.058E-06 | 3.476E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.064E-06 | 5.933E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.289E-06 | 5.851E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.374E-06 | 5.461E-04 |
|---|
| negative regulation of leukocyte proliferation | GO:0070664 |  | 2.374E-06 | 4.964E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.557E-07 | 2.87E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.622E-07 | 4.259E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 3.362E-06 | 2.065E-03 |
|---|
| ectoderm development | GO:0007398 |  | 5.405E-06 | 2.491E-03 |
|---|
| ER overload response | GO:0006983 |  | 6.69E-06 | 2.466E-03 |
|---|
| negative regulation of T cell proliferation | GO:0042130 |  | 6.69E-06 | 2.055E-03 |
|---|
| negative regulation of lymphocyte proliferation | GO:0050672 |  | 1.165E-05 | 3.067E-03 |
|---|
| negative regulation of T cell activation | GO:0050868 |  | 1.165E-05 | 2.684E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.574E-05 | 5.271E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.026E-05 | 5.577E-03 |
|---|
| regulation of protein transport | GO:0051223 |  | 3.301E-05 | 5.531E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.303E-09 | 2.997E-06 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.251E-08 | 2.589E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.262E-07 | 9.676E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.393E-07 | 1.376E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.748E-07 | 1.724E-04 |
|---|
| negative regulation of mast cell proliferation | GO:0070667 |  | 5.859E-07 | 2.246E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.058E-06 | 3.476E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.064E-06 | 5.933E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.289E-06 | 5.851E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.374E-06 | 5.461E-04 |
|---|
| negative regulation of leukocyte proliferation | GO:0070664 |  | 2.374E-06 | 4.964E-04 |
|---|



