Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6541
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7547 | 3.462e-03 | 5.610e-04 | 7.685e-01 | 1.493e-06 |
|---|
| Loi | 0.2305 | 8.000e-02 | 1.129e-02 | 4.276e-01 | 3.861e-04 |
|---|
| Schmidt | 0.6728 | 0.000e+00 | 0.000e+00 | 4.556e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.471e-03 | 0.000e+00 |
|---|
| Wang | 0.2616 | 2.679e-03 | 4.383e-02 | 3.964e-01 | 4.654e-05 |
|---|
Expression data for subnetwork 6541 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
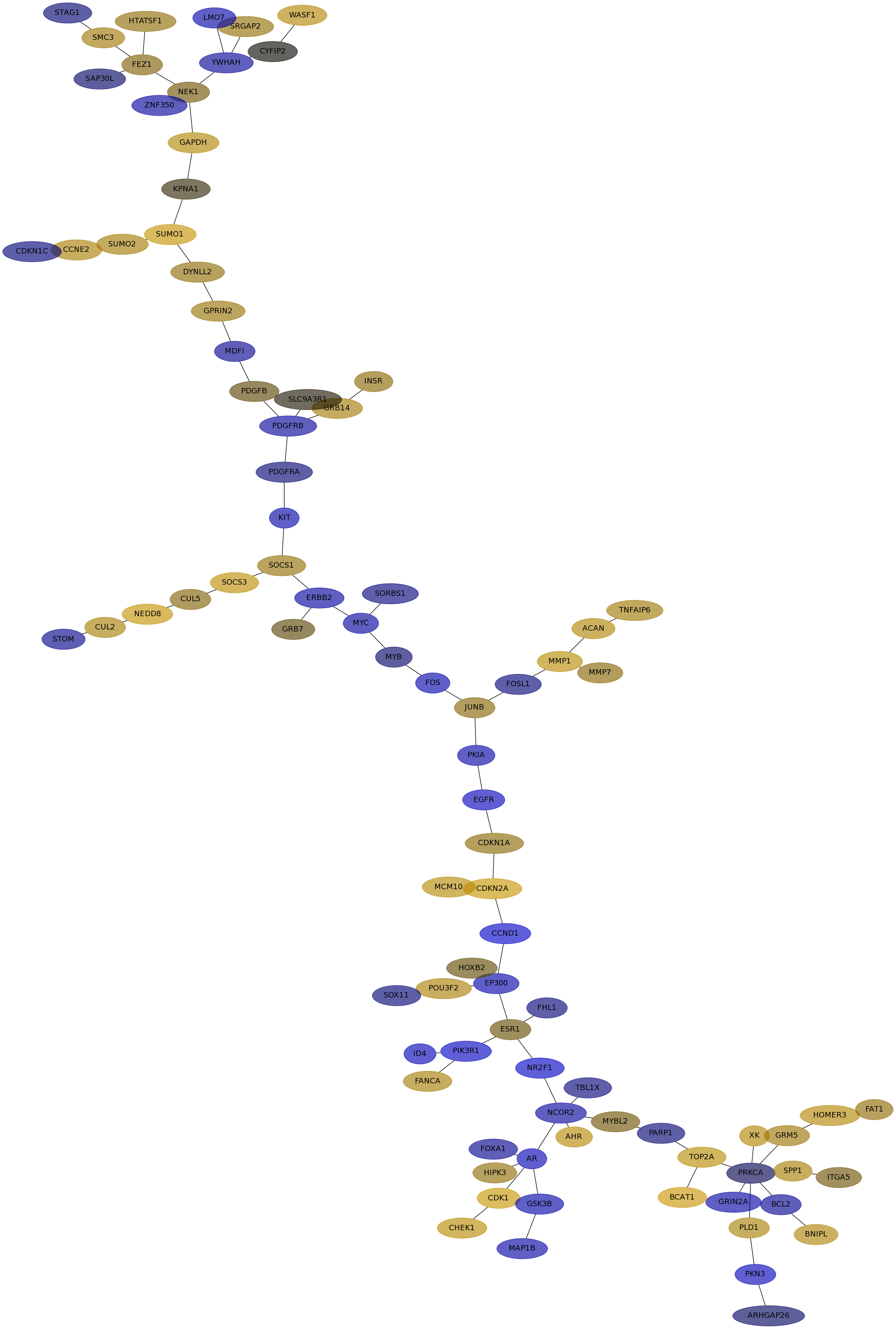
Score for each gene in subnetwork 6541 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| CCNE2 |   | 11 | 148 | 412 | 407 | 0.136 | -0.178 | 0.241 | undef | -0.055 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| LMO7 |   | 2 | 743 | 1735 | 1721 | -0.129 | 0.068 | 0.000 | 0.197 | 0.151 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| WASF1 |   | 7 | 256 | 1 | 47 | 0.162 | -0.070 | 0.348 | -0.037 | 0.130 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| BCAT1 |   | 4 | 440 | 1735 | 1699 | 0.273 | -0.134 | 0.161 | -0.078 | 0.122 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| PDGFB |   | 7 | 256 | 1165 | 1135 | 0.020 | 0.046 | 0.085 | -0.049 | 0.338 |
|---|
| PDGFRA |   | 10 | 167 | 141 | 146 | -0.035 | 0.008 | 0.118 | 0.107 | 0.119 |
|---|
| GRM5 |   | 15 | 111 | 179 | 191 | 0.094 | -0.019 | 0.000 | 0.093 | -0.093 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| MMP7 |   | 1 | 1195 | 1735 | 1746 | 0.058 | 0.066 | 0.106 | undef | 0.009 |
|---|
| SOCS1 |   | 3 | 557 | 1717 | 1689 | 0.074 | -0.153 | -0.177 | -0.202 | -0.100 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| SAP30L |   | 1 | 1195 | 1735 | 1746 | -0.025 | 0.193 | 0.057 | undef | 0.051 |
|---|
| SOX11 |   | 5 | 360 | 1396 | 1378 | -0.039 | -0.073 | -0.028 | 0.274 | -0.104 |
|---|
| PDGFRB |   | 12 | 140 | 141 | 142 | -0.094 | 0.224 | 0.127 | 0.153 | 0.189 |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| MMP1 |   | 11 | 148 | 366 | 360 | 0.192 | 0.014 | 0.036 | 0.044 | 0.260 |
|---|
| FEZ1 |   | 8 | 222 | 141 | 149 | 0.047 | -0.020 | 0.154 | 0.165 | -0.128 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| BNIPL |   | 2 | 743 | 1735 | 1721 | 0.144 | 0.072 | undef | undef | undef |
|---|
| SOCS3 |   | 2 | 743 | 1717 | 1701 | 0.201 | -0.110 | 0.064 | 0.023 | -0.138 |
|---|
| NEDD8 |   | 4 | 440 | 1650 | 1612 | 0.250 | -0.085 | 0.251 | 0.211 | -0.073 |
|---|
| HTATSF1 |   | 1 | 1195 | 1735 | 1746 | 0.071 | 0.008 | 0.247 | -0.090 | 0.070 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| FOSL1 |   | 11 | 148 | 366 | 360 | -0.040 | 0.164 | 0.010 | 0.128 | 0.005 |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| MCM10 |   | 5 | 360 | 179 | 208 | 0.178 | -0.245 | 0.015 | undef | -0.063 |
|---|
| CDKN1C |   | 1 | 1195 | 1735 | 1746 | -0.041 | 0.147 | 0.019 | 0.038 | 0.074 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| SUMO1 |   | 5 | 360 | 1034 | 1004 | 0.245 | -0.007 | 0.144 | 0.083 | 0.118 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| CUL5 |   | 3 | 557 | 1735 | 1718 | 0.051 | 0.187 | 0.221 | 0.017 | 0.216 |
|---|
| SMC3 |   | 5 | 360 | 614 | 612 | 0.105 | -0.096 | 0.238 | 0.137 | 0.047 |
|---|
| SLC9A3R1 |   | 3 | 557 | 1735 | 1718 | 0.004 | 0.017 | 0.017 | 0.096 | -0.110 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| GRIN2A |   | 12 | 140 | 179 | 192 | -0.113 | -0.068 | 0.162 | 0.125 | undef |
|---|
| ZNF350 |   | 1 | 1195 | 1735 | 1746 | -0.100 | 0.055 | 0.019 | undef | -0.084 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| SRGAP2 |   | 1 | 1195 | 1735 | 1746 | 0.078 | 0.100 | 0.261 | 0.036 | 0.152 |
|---|
| TNFAIP6 |   | 3 | 557 | 1735 | 1718 | 0.102 | 0.234 | 0.077 | -0.002 | -0.073 |
|---|
| STAG1 |   | 3 | 557 | 991 | 982 | -0.035 | 0.153 | 0.310 | 0.048 | 0.208 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| MYBL2 |   | 4 | 440 | 1604 | 1572 | 0.031 | -0.220 | 0.034 | -0.034 | 0.006 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| CYFIP2 |   | 1 | 1195 | 1735 | 1746 | 0.001 | 0.062 | -0.039 | undef | 0.126 |
|---|
| FANCA |   | 9 | 196 | 1 | 44 | 0.120 | -0.188 | 0.106 | -0.127 | -0.053 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| HOMER3 |   | 2 | 743 | 525 | 547 | 0.160 | 0.124 | -0.083 | 0.072 | 0.094 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| KPNA1 |   | 9 | 196 | 141 | 147 | 0.007 | 0.152 | -0.083 | 0.073 | 0.086 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| FAT1 |   | 2 | 743 | 1735 | 1721 | 0.071 | 0.269 | 0.087 | 0.186 | 0.094 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| CUL2 |   | 7 | 256 | 842 | 816 | 0.124 | -0.192 | -0.090 | -0.035 | 0.166 |
|---|
| ACAN |   | 5 | 360 | 412 | 416 | 0.156 | 0.132 | -0.024 | -0.171 | -0.083 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| DYNLL2 |   | 2 | 743 | 1735 | 1721 | 0.068 | -0.127 | undef | undef | undef |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| TBL1X |   | 4 | 440 | 1735 | 1699 | -0.044 | 0.017 | -0.192 | 0.198 | -0.009 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| ESR1 |   | 4 | 440 | 179 | 209 | 0.026 | -0.002 | -0.053 | -0.149 | -0.062 |
|---|
| YWHAH |   | 2 | 743 | 1323 | 1313 | -0.082 | -0.046 | 0.001 | -0.094 | -0.008 |
|---|
| AHR |   | 8 | 222 | 478 | 461 | 0.180 | 0.067 | 0.091 | -0.182 | -0.129 |
|---|
| HIPK3 |   | 9 | 196 | 1461 | 1417 | 0.069 | 0.005 | -0.027 | undef | 0.175 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| PKIA |   | 11 | 148 | 780 | 759 | -0.120 | 0.085 | 0.249 | 0.164 | 0.044 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
| STOM |   | 1 | 1195 | 1735 | 1746 | -0.066 | 0.023 | 0.223 | -0.143 | 0.085 |
|---|
GO Enrichment output for subnetwork 6541 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.081E-13 | 2.64E-10 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.358E-13 | 5.324E-10 |
|---|
| protein sumoylation | GO:0016925 |  | 1.609E-11 | 1.311E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.414E-10 | 8.633E-08 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 2.515E-10 | 1.229E-07 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 2.97E-09 | 1.209E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 9.546E-09 | 3.332E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.615E-08 | 4.932E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.85E-08 | 5.023E-06 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.14E-07 | 2.784E-05 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 1.293E-07 | 2.871E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.705E-13 | 4.103E-10 |
|---|
| interphase | GO:0051325 |  | 2.578E-13 | 3.101E-10 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.663E-13 | 6.948E-10 |
|---|
| protein sumoylation | GO:0016925 |  | 2.461E-11 | 1.48E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.612E-10 | 1.257E-07 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 3.588E-10 | 1.439E-07 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 4.232E-09 | 1.455E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.451E-08 | 4.365E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.453E-08 | 6.558E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.991E-08 | 7.197E-06 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.621E-07 | 3.547E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.051E-12 | 1.622E-08 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.218E-11 | 3.7E-08 |
|---|
| interphase | GO:0051325 |  | 4.251E-11 | 3.259E-08 |
|---|
| protein sumoylation | GO:0016925 |  | 1.413E-10 | 8.125E-08 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.549E-09 | 7.126E-07 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 9.152E-09 | 3.508E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.632E-08 | 8.647E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.128E-08 | 1.762E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.151E-08 | 2.339E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.058E-07 | 2.434E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 5.203E-07 | 1.088E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.32E-08 | 4.276E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 4.185E-08 | 3.857E-05 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.351E-07 | 8.298E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.179E-07 | 1.004E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.895E-07 | 1.067E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 7.285E-07 | 2.238E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 8.065E-07 | 2.123E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.188E-07 | 2.117E-04 |
|---|
| regulation of protein amino acid phosphorylation | GO:0001932 |  | 1.017E-06 | 2.083E-04 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 1.206E-06 | 2.223E-04 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 1.753E-06 | 2.937E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.051E-12 | 1.622E-08 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.218E-11 | 3.7E-08 |
|---|
| interphase | GO:0051325 |  | 4.251E-11 | 3.259E-08 |
|---|
| protein sumoylation | GO:0016925 |  | 1.413E-10 | 8.125E-08 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.549E-09 | 7.126E-07 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 9.152E-09 | 3.508E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.632E-08 | 8.647E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.128E-08 | 1.762E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.151E-08 | 2.339E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.058E-07 | 2.434E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 5.203E-07 | 1.088E-04 |
|---|



