Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6502
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7552 | 3.432e-03 | 5.550e-04 | 7.672e-01 | 1.461e-06 |
|---|
| Loi | 0.2302 | 8.050e-02 | 1.142e-02 | 4.288e-01 | 3.942e-04 |
|---|
| Schmidt | 0.6725 | 0.000e+00 | 0.000e+00 | 4.591e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.474e-03 | 0.000e+00 |
|---|
| Wang | 0.2613 | 2.714e-03 | 4.415e-02 | 3.979e-01 | 4.767e-05 |
|---|
Expression data for subnetwork 6502 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
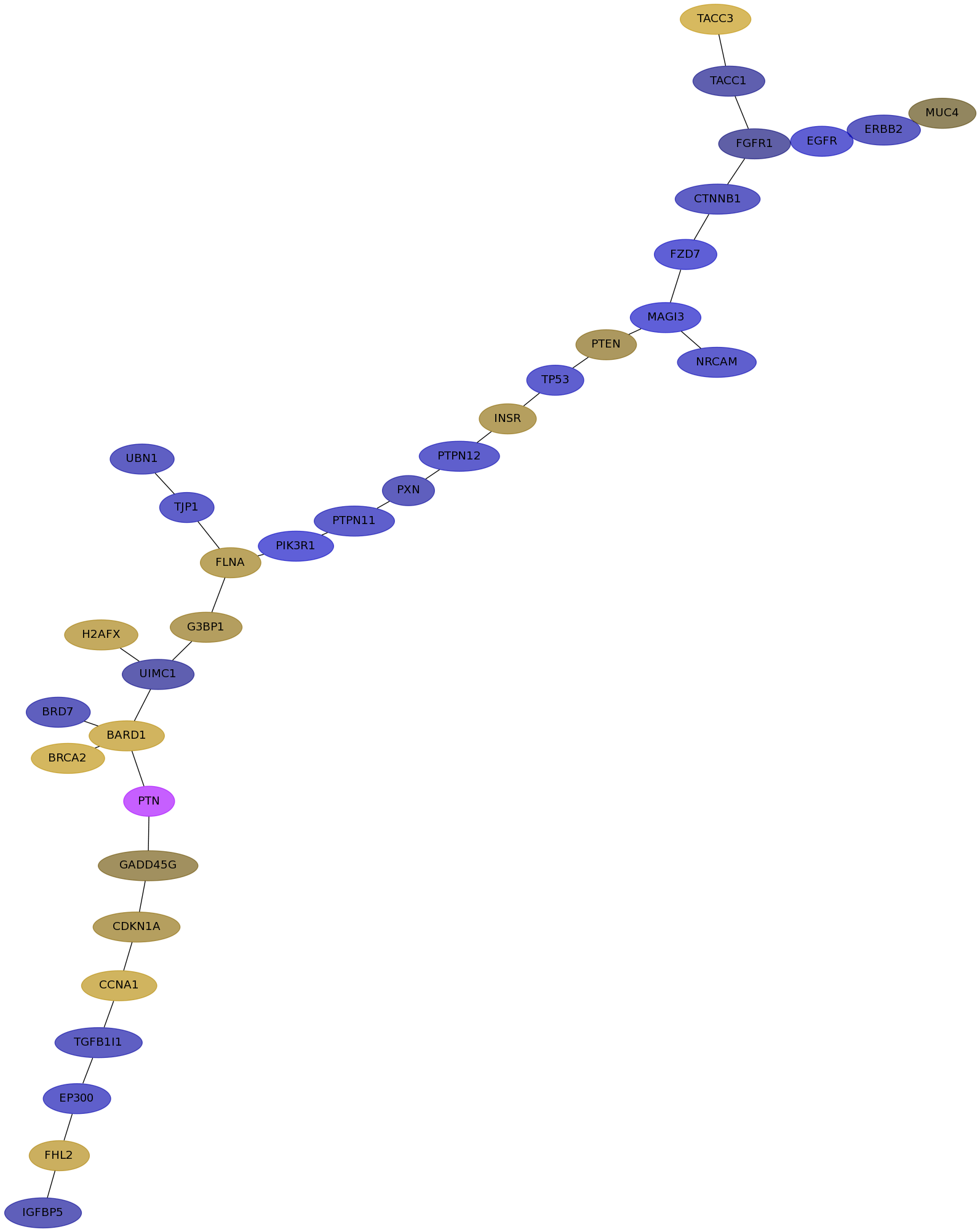
Score for each gene in subnetwork 6502 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| UBN1 |   | 3 | 557 | 648 | 647 | -0.106 | 0.059 | 0.130 | -0.144 | 0.066 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| TJP1 |   | 8 | 222 | 318 | 325 | -0.139 | 0.102 | 0.099 | 0.319 | 0.049 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| NRCAM |   | 2 | 743 | 1842 | 1829 | -0.163 | 0.111 | 0.112 | 0.090 | -0.138 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| MUC4 |   | 1 | 1195 | 1842 | 1855 | 0.017 | 0.135 | -0.097 | undef | -0.076 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| TACC3 |   | 4 | 440 | 1077 | 1065 | 0.232 | -0.144 | 0.014 | 0.046 | 0.004 |
|---|
| PXN |   | 12 | 140 | 1 | 36 | -0.088 | 0.284 | -0.022 | 0.060 | 0.114 |
|---|
| TACC1 |   | 5 | 360 | 1077 | 1057 | -0.051 | 0.020 | -0.022 | 0.187 | 0.126 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| PTPN12 |   | 6 | 301 | 478 | 473 | -0.159 | 0.153 | 0.031 | 0.083 | 0.042 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| MAGI3 |   | 2 | 743 | 1842 | 1829 | -0.233 | 0.199 | undef | undef | undef |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| FZD7 |   | 2 | 743 | 1396 | 1391 | -0.219 | 0.062 | 0.062 | -0.110 | -0.059 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| H2AFX |   | 7 | 256 | 179 | 201 | 0.111 | -0.072 | 0.121 | 0.165 | -0.010 |
|---|
| UIMC1 |   | 6 | 301 | 179 | 205 | -0.054 | 0.226 | 0.031 | -0.005 | 0.189 |
|---|
| G3BP1 |   | 7 | 256 | 1116 | 1082 | 0.060 | -0.077 | -0.017 | 0.155 | -0.004 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
GO Enrichment output for subnetwork 6502 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 4.067E-12 | 9.936E-09 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.709E-11 | 4.53E-08 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 7.288E-11 | 5.935E-08 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 3.45E-10 | 2.107E-07 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 3.507E-10 | 1.713E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.033E-09 | 4.204E-07 |
|---|
| organ growth | GO:0035265 |  | 2.404E-09 | 8.39E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 2.404E-09 | 7.341E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 2.404E-09 | 6.525E-07 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.797E-09 | 1.172E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 4.797E-09 | 1.065E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.187E-12 | 1.489E-08 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 6.591E-11 | 7.929E-08 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.294E-10 | 1.038E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 4.835E-10 | 2.908E-07 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 6.213E-10 | 2.99E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.447E-09 | 5.802E-07 |
|---|
| organ growth | GO:0035265 |  | 3.368E-09 | 1.158E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 3.368E-09 | 1.013E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 3.368E-09 | 9.003E-07 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.719E-09 | 1.617E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 6.719E-09 | 1.47E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 4.943E-12 | 1.137E-08 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 6.724E-11 | 7.733E-08 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.64E-10 | 2.024E-07 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 8.224E-10 | 4.729E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.108E-09 | 5.096E-07 |
|---|
| organ growth | GO:0035265 |  | 7.708E-09 | 2.955E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 7.708E-09 | 2.533E-06 |
|---|
| negative regulation of transport | GO:0051051 |  | 1.448E-08 | 4.162E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 1.537E-08 | 3.928E-06 |
|---|
| regulation of protein transport | GO:0051223 |  | 1.924E-08 | 4.426E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 4.225E-08 | 8.834E-06 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.096E-08 | 3.864E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 3.17E-08 | 2.921E-05 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 6.607E-08 | 4.059E-05 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 4.683E-07 | 2.158E-04 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 5.372E-07 | 1.98E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 7.097E-07 | 2.18E-04 |
|---|
| negative regulation of protein transport | GO:0051224 |  | 7.786E-07 | 2.05E-04 |
|---|
| negative regulation of transport | GO:0051051 |  | 7.867E-07 | 1.812E-04 |
|---|
| regulation of protein transport | GO:0051223 |  | 1.058E-06 | 2.166E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.372E-06 | 2.529E-04 |
|---|
| regulation of protein localization | GO:0032880 |  | 1.528E-06 | 2.561E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 4.943E-12 | 1.137E-08 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 6.724E-11 | 7.733E-08 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.64E-10 | 2.024E-07 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 8.224E-10 | 4.729E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.108E-09 | 5.096E-07 |
|---|
| organ growth | GO:0035265 |  | 7.708E-09 | 2.955E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 7.708E-09 | 2.533E-06 |
|---|
| negative regulation of transport | GO:0051051 |  | 1.448E-08 | 4.162E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 1.537E-08 | 3.928E-06 |
|---|
| regulation of protein transport | GO:0051223 |  | 1.924E-08 | 4.426E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 4.225E-08 | 8.834E-06 |
|---|



