Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6483
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7552 | 3.434e-03 | 5.550e-04 | 7.673e-01 | 1.462e-06 |
|---|
| Loi | 0.2302 | 8.046e-02 | 1.141e-02 | 4.287e-01 | 3.936e-04 |
|---|
| Schmidt | 0.6724 | 0.000e+00 | 0.000e+00 | 4.602e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.465e-03 | 0.000e+00 |
|---|
| Wang | 0.2611 | 2.736e-03 | 4.434e-02 | 3.988e-01 | 4.838e-05 |
|---|
Expression data for subnetwork 6483 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
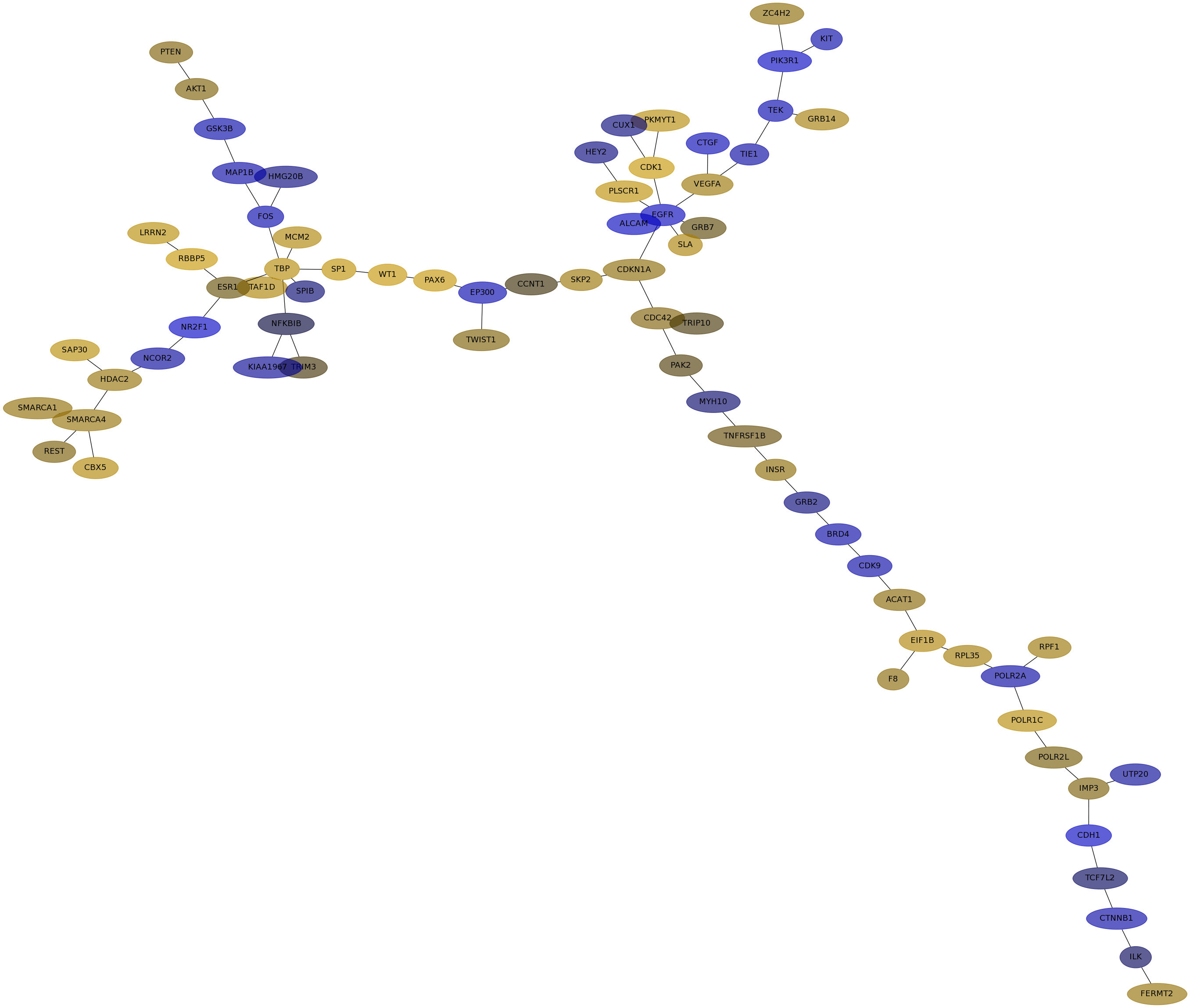
Score for each gene in subnetwork 6483 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| RPF1 |   | 1 | 1195 | 1938 | 1946 | 0.089 | -0.113 | 0.059 | undef | 0.076 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| TRIP10 |   | 5 | 360 | 1034 | 1004 | 0.012 | 0.154 | 0.049 | undef | 0.144 |
|---|
| SMARCA4 |   | 4 | 440 | 1364 | 1330 | 0.080 | 0.202 | -0.071 | -0.050 | -0.116 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| MYH10 |   | 5 | 360 | 236 | 242 | -0.029 | 0.139 | 0.132 | undef | 0.223 |
|---|
| EIF1B |   | 8 | 222 | 1576 | 1529 | 0.149 | 0.075 | 0.332 | -0.017 | -0.070 |
|---|
| HEY2 |   | 8 | 222 | 570 | 555 | -0.045 | -0.021 | 0.398 | -0.024 | -0.058 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| REST |   | 1 | 1195 | 1938 | 1946 | 0.040 | -0.014 | 0.067 | 0.029 | 0.099 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| SAP30 |   | 2 | 743 | 1938 | 1922 | 0.186 | 0.083 | 0.153 | -0.047 | -0.184 |
|---|
| BRD4 |   | 11 | 148 | 1047 | 1003 | -0.119 | 0.095 | -0.075 | 0.081 | -0.044 |
|---|
| RPL35 |   | 8 | 222 | 1323 | 1280 | 0.108 | -0.012 | 0.370 | -0.145 | -0.220 |
|---|
| POLR2L |   | 5 | 360 | 727 | 722 | 0.037 | -0.012 | 0.224 | 0.054 | 0.015 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| SMARCA1 |   | 1 | 1195 | 1938 | 1946 | 0.070 | 0.059 | 0.146 | -0.029 | 0.044 |
|---|
| CUX1 |   | 6 | 301 | 1203 | 1169 | -0.042 | 0.256 | -0.121 | 0.105 | 0.040 |
|---|
| KIAA1967 |   | 1 | 1195 | 1938 | 1946 | -0.077 | 0.075 | 0.162 | -0.121 | undef |
|---|
| PKMYT1 |   | 7 | 256 | 1510 | 1463 | 0.175 | -0.143 | 0.046 | 0.105 | -0.070 |
|---|
| MCM2 |   | 2 | 743 | 1846 | 1832 | 0.148 | -0.200 | 0.074 | -0.045 | 0.168 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| ALCAM |   | 8 | 222 | 1 | 46 | -0.208 | 0.193 | 0.067 | 0.088 | -0.010 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| CCNT1 |   | 6 | 301 | 478 | 473 | 0.008 | -0.028 | 0.162 | 0.007 | -0.213 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| HDAC2 |   | 13 | 124 | 318 | 316 | 0.079 | -0.094 | 0.148 | 0.089 | 0.037 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| TBP |   | 4 | 440 | 1141 | 1127 | 0.171 | 0.017 | 0.009 | -0.061 | -0.009 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| F8 |   | 1 | 1195 | 1938 | 1946 | 0.059 | -0.037 | 0.088 | -0.042 | 0.039 |
|---|
| SLA |   | 2 | 743 | 1938 | 1922 | 0.138 | -0.195 | -0.058 | undef | -0.012 |
|---|
| SPIB |   | 2 | 743 | 1803 | 1782 | -0.034 | -0.202 | -0.070 | -0.102 | -0.179 |
|---|
| RBBP5 |   | 3 | 557 | 478 | 487 | 0.264 | 0.053 | 0.086 | undef | 0.033 |
|---|
| LRRN2 |   | 1 | 1195 | 1938 | 1946 | 0.189 | 0.061 | -0.057 | 0.059 | 0.061 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| TRIM3 |   | 2 | 743 | 1116 | 1121 | 0.010 | 0.009 | -0.118 | undef | 0.063 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PLSCR1 |   | 10 | 167 | 412 | 408 | 0.202 | -0.159 | 0.091 | undef | -0.105 |
|---|
| TAF1D |   | 10 | 167 | 590 | 566 | 0.141 | 0.113 | 0.041 | undef | 0.059 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| HMG20B |   | 2 | 743 | 1893 | 1869 | -0.045 | 0.146 | -0.151 | -0.074 | -0.155 |
|---|
| FERMT2 |   | 2 | 743 | 1413 | 1410 | 0.077 | 0.151 | 0.219 | undef | 0.287 |
|---|
| ILK |   | 4 | 440 | 1413 | 1390 | -0.020 | 0.195 | 0.076 | 0.130 | 0.086 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| IMP3 |   | 4 | 440 | 366 | 381 | 0.042 | -0.132 | 0.113 | -0.304 | -0.196 |
|---|
| POLR2A |   | 6 | 301 | 1547 | 1510 | -0.107 | 0.247 | 0.091 | 0.040 | 0.141 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| TNFRSF1B |   | 2 | 743 | 1938 | 1922 | 0.024 | -0.187 | -0.096 | -0.228 | -0.106 |
|---|
| UTP20 |   | 1 | 1195 | 1938 | 1946 | -0.082 | -0.162 | 0.039 | 0.109 | -0.148 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CDK9 |   | 3 | 557 | 1598 | 1574 | -0.120 | 0.179 | 0.182 | 0.186 | -0.135 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| ACAT1 |   | 2 | 743 | 1 | 66 | 0.057 | -0.011 | 0.087 | 0.038 | 0.112 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| POLR1C |   | 3 | 557 | 1323 | 1309 | 0.176 | -0.210 | 0.181 | -0.126 | 0.038 |
|---|
| NFKBIB |   | 2 | 743 | 1938 | 1922 | -0.008 | -0.192 | -0.185 | -0.098 | -0.239 |
|---|
| CBX5 |   | 5 | 360 | 1 | 56 | 0.155 | 0.016 | 0.148 | 0.030 | -0.009 |
|---|
| ESR1 |   | 4 | 440 | 179 | 209 | 0.026 | -0.002 | -0.053 | -0.149 | -0.062 |
|---|
GO Enrichment output for subnetwork 6483 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.738E-12 | 2.379E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.124E-11 | 2.594E-08 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 3.939E-09 | 3.208E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.177E-08 | 7.187E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.432E-08 | 1.188E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.432E-08 | 9.901E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 2.734E-08 | 9.542E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.445E-08 | 1.663E-05 |
|---|
| lung development | GO:0030324 |  | 9.103E-08 | 2.471E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 9.761E-08 | 2.385E-05 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 1.013E-07 | 2.249E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 7.274E-12 | 1.75E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.347E-11 | 5.229E-08 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 5.483E-09 | 4.397E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.637E-08 | 9.849E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.594E-08 | 1.248E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.658E-08 | 1.467E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.803E-08 | 1.307E-05 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 3.803E-08 | 1.144E-05 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 7.572E-08 | 2.024E-05 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 9.036E-08 | 2.174E-05 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 1.18E-07 | 2.582E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.239E-09 | 1.205E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 5.193E-08 | 5.972E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.394E-07 | 1.836E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 7.097E-07 | 4.081E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.988E-07 | 4.595E-04 |
|---|
| nuclear migration | GO:0007097 |  | 4.553E-06 | 1.745E-03 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 4.553E-06 | 1.496E-03 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 4.553E-06 | 1.309E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.553E-06 | 1.164E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 6.143E-06 | 1.413E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 7.709E-06 | 1.612E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell migration | GO:0030334 |  | 1.111E-06 | 2.048E-03 |
|---|
| regulation of locomotion | GO:0040012 |  | 2.308E-06 | 2.127E-03 |
|---|
| regulation of cell motion | GO:0051270 |  | 2.308E-06 | 1.418E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.695E-06 | 3.085E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 7.728E-06 | 2.849E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 9.562E-06 | 2.937E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.017E-05 | 2.677E-03 |
|---|
| regulation of RNA stability | GO:0043487 |  | 2.49E-05 | 5.736E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.648E-05 | 5.422E-03 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.714E-05 | 6.845E-03 |
|---|
| transcription initiation | GO:0006352 |  | 3.997E-05 | 6.696E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.239E-09 | 1.205E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 5.193E-08 | 5.972E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.394E-07 | 1.836E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 7.097E-07 | 4.081E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.988E-07 | 4.595E-04 |
|---|
| nuclear migration | GO:0007097 |  | 4.553E-06 | 1.745E-03 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 4.553E-06 | 1.496E-03 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 4.553E-06 | 1.309E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.553E-06 | 1.164E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 6.143E-06 | 1.413E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 7.709E-06 | 1.612E-03 |
|---|



