Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6481
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7553 | 3.426e-03 | 5.530e-04 | 7.669e-01 | 1.453e-06 |
|---|
| Loi | 0.2301 | 8.062e-02 | 1.145e-02 | 4.291e-01 | 3.961e-04 |
|---|
| Schmidt | 0.6724 | 0.000e+00 | 0.000e+00 | 4.602e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.462e-03 | 0.000e+00 |
|---|
| Wang | 0.2611 | 2.738e-03 | 4.436e-02 | 3.989e-01 | 4.845e-05 |
|---|
Expression data for subnetwork 6481 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
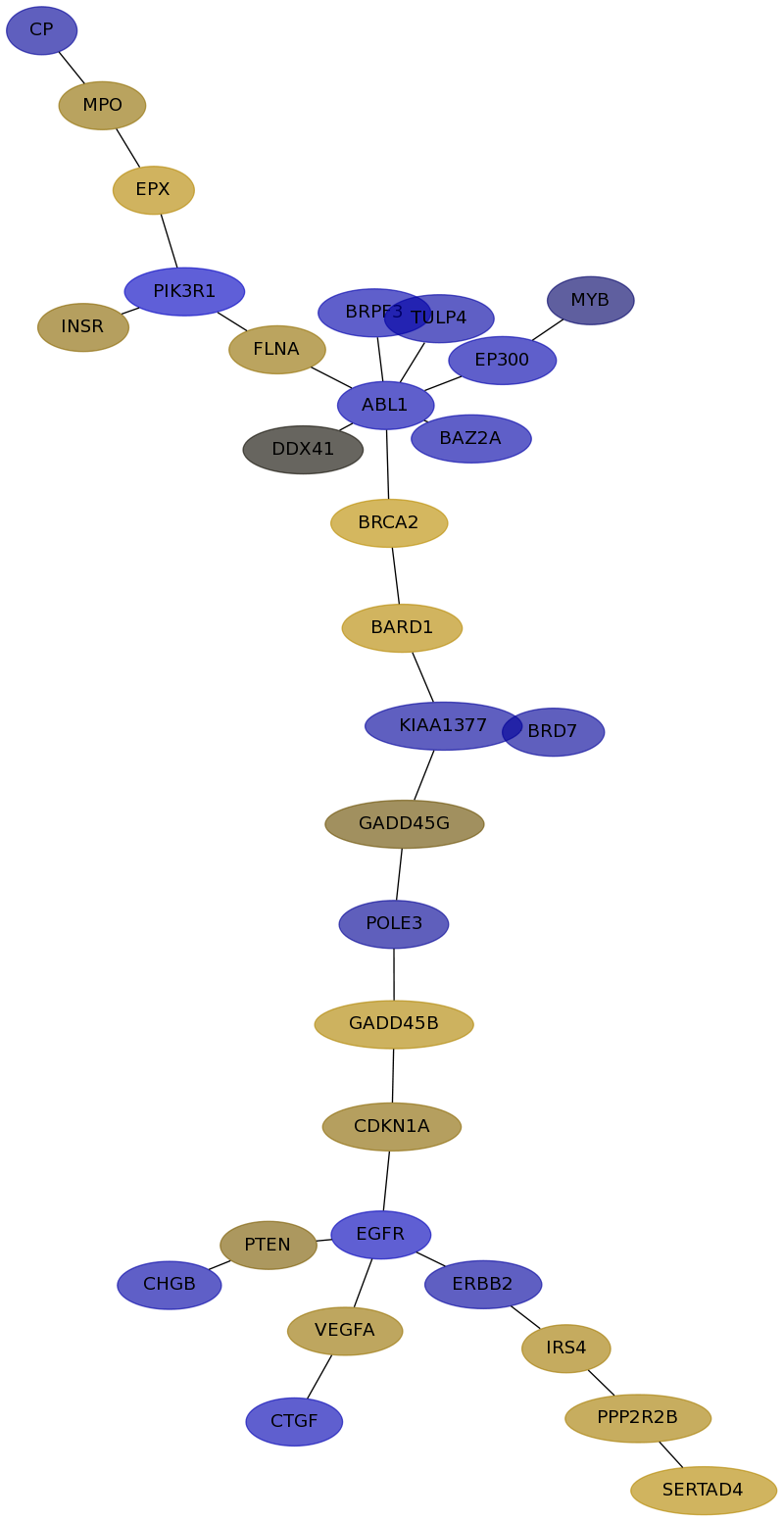
Score for each gene in subnetwork 6481 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| CHGB |   | 7 | 256 | 682 | 664 | -0.126 | -0.041 | 0.018 | 0.102 | -0.131 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| CP |   | 1 | 1195 | 1889 | 1898 | -0.090 | 0.106 | -0.015 | 0.237 | 0.061 |
|---|
| DDX41 |   | 1 | 1195 | 1889 | 1898 | 0.002 | 0.154 | -0.126 | 0.251 | 0.184 |
|---|
| BAZ2A |   | 1 | 1195 | 1889 | 1898 | -0.127 | 0.029 | -0.033 | 0.182 | -0.097 |
|---|
| SERTAD4 |   | 8 | 222 | 1268 | 1225 | 0.173 | 0.169 | undef | -0.050 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| EPX |   | 3 | 557 | 478 | 487 | 0.172 | -0.224 | -0.005 | 0.173 | -0.064 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| MPO |   | 1 | 1195 | 1889 | 1898 | 0.074 | -0.006 | 0.000 | -0.002 | 0.079 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| BRPF3 |   | 2 | 743 | 806 | 806 | -0.145 | 0.028 | undef | 0.218 | undef |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| GADD45B |   | 8 | 222 | 318 | 325 | 0.158 | 0.122 | 0.067 | 0.060 | 0.212 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| TULP4 |   | 2 | 743 | 1358 | 1360 | -0.114 | 0.067 | 0.024 | -0.099 | 0.047 |
|---|
GO Enrichment output for subnetwork 6481 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 8.639E-11 | 2.11E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.068E-10 | 2.527E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.587E-10 | 2.107E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 6.028E-10 | 3.682E-07 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.204E-09 | 5.881E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.208E-09 | 8.992E-07 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 3.6E-09 | 1.256E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 5.648E-09 | 1.725E-06 |
|---|
| regulation of protein stability | GO:0031647 |  | 1.622E-08 | 4.402E-06 |
|---|
| copper ion transport | GO:0006825 |  | 2.321E-08 | 5.671E-06 |
|---|
| response to nutrient | GO:0007584 |  | 2.367E-08 | 5.256E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.335E-10 | 3.213E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.551E-10 | 4.272E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.999E-10 | 3.207E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 9.314E-10 | 5.603E-07 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.86E-09 | 8.948E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.185E-09 | 1.678E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 5.559E-09 | 1.911E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 8.721E-09 | 2.623E-06 |
|---|
| regulation of protein stability | GO:0031647 |  | 2.413E-08 | 6.45E-06 |
|---|
| copper ion transport | GO:0006825 |  | 3.582E-08 | 8.618E-06 |
|---|
| response to nutrient | GO:0007584 |  | 3.859E-08 | 8.44E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.227E-08 | 2.823E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.193E-07 | 2.521E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.316E-07 | 2.542E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 4.376E-07 | 2.516E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 4.376E-07 | 2.013E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 7.643E-07 | 2.93E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.22E-06 | 4.01E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.22E-06 | 3.509E-04 |
|---|
| regulation of protein stability | GO:0031647 |  | 2.007E-06 | 5.128E-04 |
|---|
| response to nutrient | GO:0007584 |  | 2.314E-06 | 5.322E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.605E-06 | 5.446E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 1.494E-07 | 2.753E-04 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 1.715E-07 | 1.58E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.253E-06 | 7.696E-04 |
|---|
| regulation of cell migration | GO:0030334 |  | 1.817E-06 | 8.37E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.035E-06 | 7.501E-04 |
|---|
| regulation of locomotion | GO:0040012 |  | 3.096E-06 | 9.509E-04 |
|---|
| regulation of cell motion | GO:0051270 |  | 3.096E-06 | 8.151E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 3.547E-06 | 8.173E-04 |
|---|
| activation of MAPKK activity | GO:0000186 |  | 3.982E-06 | 8.154E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.982E-06 | 7.339E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 4.924E-06 | 8.25E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.227E-08 | 2.823E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.193E-07 | 2.521E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.316E-07 | 2.542E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 4.376E-07 | 2.516E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 4.376E-07 | 2.013E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 7.643E-07 | 2.93E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.22E-06 | 4.01E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.22E-06 | 3.509E-04 |
|---|
| regulation of protein stability | GO:0031647 |  | 2.007E-06 | 5.128E-04 |
|---|
| response to nutrient | GO:0007584 |  | 2.314E-06 | 5.322E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.605E-06 | 5.446E-04 |
|---|



