Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6474
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7553 | 3.424e-03 | 5.530e-04 | 7.669e-01 | 1.452e-06 |
|---|
| Loi | 0.2300 | 8.070e-02 | 1.147e-02 | 4.293e-01 | 3.974e-04 |
|---|
| Schmidt | 0.6725 | 0.000e+00 | 0.000e+00 | 4.594e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.466e-03 | 0.000e+00 |
|---|
| Wang | 0.2612 | 2.734e-03 | 4.433e-02 | 3.987e-01 | 4.832e-05 |
|---|
Expression data for subnetwork 6474 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
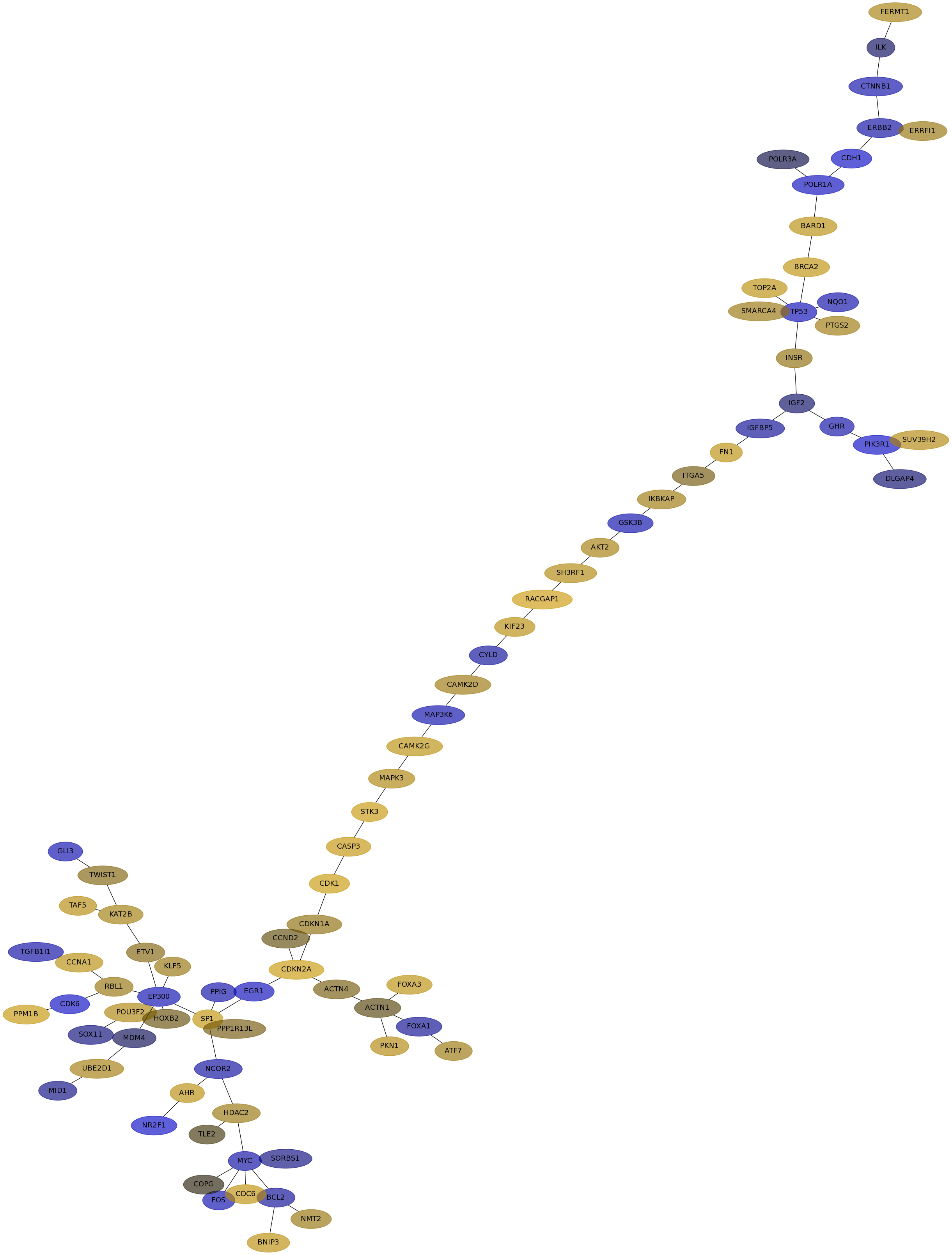
Score for each gene in subnetwork 6474 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| CASP3 |   | 11 | 148 | 141 | 143 | 0.234 | -0.032 | -0.142 | -0.097 | -0.084 |
|---|
| CYLD |   | 1 | 1195 | 1873 | 1882 | -0.083 | -0.137 | -0.250 | undef | -0.011 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| POLR1A |   | 6 | 301 | 457 | 452 | -0.193 | -0.066 | undef | 0.093 | undef |
|---|
| MDM4 |   | 9 | 196 | 141 | 147 | -0.016 | 0.004 | 0.164 | 0.107 | 0.105 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| SMARCA4 |   | 4 | 440 | 1364 | 1330 | 0.080 | 0.202 | -0.071 | -0.050 | -0.116 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| PPM1B |   | 1 | 1195 | 1873 | 1882 | 0.241 | -0.038 | -0.019 | -0.124 | -0.045 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| UBE2D1 |   | 1 | 1195 | 1873 | 1882 | 0.105 | 0.045 | 0.121 | 0.056 | -0.066 |
|---|
| ETV1 |   | 10 | 167 | 1 | 40 | 0.046 | 0.054 | -0.065 | 0.014 | 0.141 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| SOX11 |   | 5 | 360 | 1396 | 1378 | -0.039 | -0.073 | -0.028 | 0.274 | -0.104 |
|---|
| TAF5 |   | 5 | 360 | 478 | 476 | 0.166 | -0.131 | 0.125 | undef | 0.076 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| CDC6 |   | 3 | 557 | 570 | 572 | 0.198 | -0.053 | 0.038 | -0.202 | 0.095 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| MAP3K6 |   | 2 | 743 | 638 | 650 | -0.129 | 0.287 | -0.015 | 0.248 | 0.066 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| FERMT1 |   | 1 | 1195 | 1873 | 1882 | 0.111 | 0.083 | -0.052 | -0.186 | -0.017 |
|---|
| PTGS2 |   | 2 | 743 | 366 | 395 | 0.091 | -0.073 | 0.250 | -0.177 | 0.152 |
|---|
| CAMK2D |   | 2 | 743 | 638 | 650 | 0.085 | -0.149 | 0.000 | undef | undef |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| SH3RF1 |   | 6 | 301 | 1222 | 1194 | 0.144 | 0.238 | undef | undef | undef |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| MID1 |   | 1 | 1195 | 1873 | 1882 | -0.047 | 0.156 | 0.068 | undef | 0.166 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| HDAC2 |   | 13 | 124 | 318 | 316 | 0.079 | -0.094 | 0.148 | 0.089 | 0.037 |
|---|
| COPG |   | 5 | 360 | 570 | 562 | 0.004 | 0.080 | -0.158 | undef | 0.280 |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| FN1 |   | 6 | 301 | 590 | 582 | 0.190 | 0.133 | 0.091 | undef | 0.083 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| DLGAP4 |   | 7 | 256 | 727 | 714 | -0.029 | 0.180 | -0.201 | 0.135 | -0.043 |
|---|
| FOXA3 |   | 12 | 140 | 318 | 322 | 0.192 | 0.289 | undef | -0.171 | undef |
|---|
| KIF23 |   | 4 | 440 | 957 | 942 | 0.168 | -0.063 | 0.162 | undef | 0.033 |
|---|
| RACGAP1 |   | 4 | 440 | 1222 | 1205 | 0.283 | -0.164 | 0.073 | 0.055 | 0.188 |
|---|
| IKBKAP |   | 10 | 167 | 412 | 408 | 0.089 | 0.184 | 0.142 | 0.105 | -0.017 |
|---|
| PKN1 |   | 5 | 360 | 1674 | 1634 | 0.150 | -0.143 | -0.011 | 0.084 | -0.002 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| SUV39H2 |   | 8 | 222 | 318 | 325 | 0.150 | -0.097 | -0.057 | undef | 0.028 |
|---|
| STK3 |   | 4 | 440 | 141 | 165 | 0.254 | 0.007 | 0.106 | 0.326 | 0.198 |
|---|
| PPIG |   | 8 | 222 | 648 | 631 | -0.116 | 0.024 | 0.221 | -0.065 | 0.101 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| BNIP3 |   | 11 | 148 | 236 | 232 | 0.193 | 0.071 | 0.102 | 0.119 | 0.285 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| CCND2 |   | 5 | 360 | 1492 | 1453 | 0.022 | -0.011 | -0.072 | -0.153 | -0.029 |
|---|
| ERRFI1 |   | 10 | 167 | 412 | 408 | 0.076 | 0.215 | undef | -0.043 | undef |
|---|
| CAMK2G |   | 1 | 1195 | 1873 | 1882 | 0.191 | -0.004 | 0.103 | 0.045 | -0.072 |
|---|
| TLE2 |   | 4 | 440 | 806 | 790 | 0.010 | 0.052 | 0.202 | undef | 0.093 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| ILK |   | 4 | 440 | 1413 | 1390 | -0.020 | 0.195 | 0.076 | 0.130 | 0.086 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| POLR3A |   | 8 | 222 | 296 | 287 | -0.010 | -0.032 | undef | 0.091 | undef |
|---|
| NMT2 |   | 7 | 256 | 906 | 876 | 0.077 | -0.296 | 0.134 | -0.124 | 0.217 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| MAPK3 |   | 6 | 301 | 141 | 152 | 0.137 | -0.102 | -0.126 | undef | -0.086 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| PPP1R13L |   | 2 | 743 | 478 | 490 | 0.032 | 0.351 | -0.104 | 0.042 | 0.173 |
|---|
| AHR |   | 8 | 222 | 478 | 461 | 0.180 | 0.067 | 0.091 | -0.182 | -0.129 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
| IGF2 |   | 14 | 117 | 570 | 553 | -0.022 | 0.172 | 0.145 | 0.087 | 0.116 |
|---|
GO Enrichment output for subnetwork 6474 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.893E-09 | 4.625E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.607E-09 | 4.406E-06 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 7.106E-09 | 5.787E-06 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.661E-08 | 1.014E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 5.055E-08 | 2.47E-05 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 6.965E-08 | 2.836E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.629E-07 | 5.686E-05 |
|---|
| nucleus organization | GO:0006997 |  | 1.691E-07 | 5.163E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.85E-07 | 5.022E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.913E-07 | 7.116E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 3.352E-07 | 7.444E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.12E-09 | 7.507E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 5.939E-09 | 7.144E-06 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 9.967E-09 | 7.994E-06 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.524E-08 | 1.518E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 7.673E-08 | 3.692E-05 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 1.057E-07 | 4.237E-05 |
|---|
| nucleus organization | GO:0006997 |  | 2.009E-07 | 6.904E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.468E-07 | 7.424E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.731E-07 | 7.3E-05 |
|---|
| interphase | GO:0051325 |  | 3.475E-07 | 8.36E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 4.077E-07 | 8.917E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.814E-09 | 1.107E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 6.988E-09 | 8.036E-06 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.194E-08 | 1.682E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 6.67E-08 | 3.835E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 9.939E-08 | 4.572E-05 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 2.021E-07 | 7.747E-05 |
|---|
| nucleus organization | GO:0006997 |  | 2.594E-07 | 8.524E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.012E-07 | 8.661E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.747E-07 | 9.577E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.381E-07 | 1.238E-04 |
|---|
| interphase | GO:0051325 |  | 6.429E-07 | 1.344E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.329E-07 | 6.135E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 5.708E-07 | 5.259E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 5.954E-07 | 3.658E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 9.86E-07 | 4.543E-04 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 9.86E-07 | 3.634E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.539E-06 | 4.729E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.539E-06 | 4.053E-04 |
|---|
| hindbrain development | GO:0030902 |  | 1.539E-06 | 3.547E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.819E-06 | 3.724E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.727E-06 | 5.026E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 3.224E-06 | 5.401E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.814E-09 | 1.107E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 6.988E-09 | 8.036E-06 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.194E-08 | 1.682E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 6.67E-08 | 3.835E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 9.939E-08 | 4.572E-05 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 2.021E-07 | 7.747E-05 |
|---|
| nucleus organization | GO:0006997 |  | 2.594E-07 | 8.524E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.012E-07 | 8.661E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.747E-07 | 9.577E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.381E-07 | 1.238E-04 |
|---|
| interphase | GO:0051325 |  | 6.429E-07 | 1.344E-04 |
|---|



