Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6473
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7554 | 3.422e-03 | 5.530e-04 | 7.668e-01 | 1.451e-06 |
|---|
| Loi | 0.2300 | 8.077e-02 | 1.149e-02 | 4.294e-01 | 3.986e-04 |
|---|
| Schmidt | 0.6725 | 0.000e+00 | 0.000e+00 | 4.591e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.465e-03 | 0.000e+00 |
|---|
| Wang | 0.2611 | 2.740e-03 | 4.438e-02 | 3.990e-01 | 4.851e-05 |
|---|
Expression data for subnetwork 6473 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
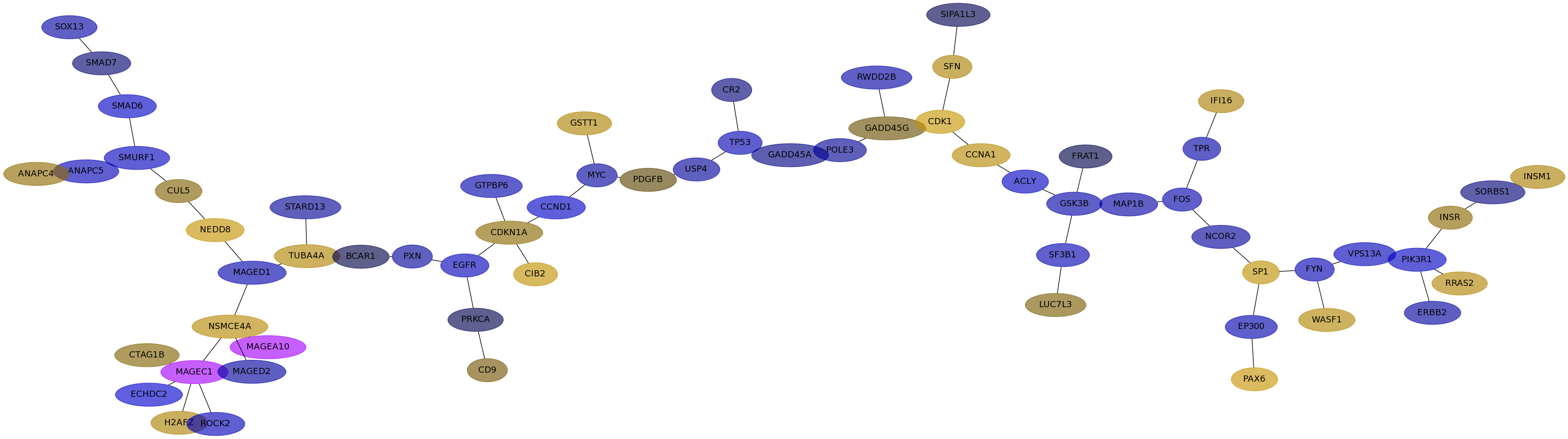
Score for each gene in subnetwork 6473 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| H2AFZ |   | 1 | 1195 | 1879 | 1889 | 0.141 | -0.058 | 0.168 | 0.231 | 0.138 |
|---|
| WASF1 |   | 7 | 256 | 1 | 47 | 0.162 | -0.070 | 0.348 | -0.037 | 0.130 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| MAGED1 |   | 5 | 360 | 842 | 828 | -0.141 | 0.284 | -0.152 | 0.240 | 0.045 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| VPS13A |   | 10 | 167 | 236 | 234 | -0.203 | 0.105 | -0.046 | 0.120 | 0.062 |
|---|
| PDGFB |   | 7 | 256 | 1165 | 1135 | 0.020 | 0.046 | 0.085 | -0.049 | 0.338 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| TUBA4A |   | 7 | 256 | 1 | 47 | 0.157 | 0.136 | -0.010 | undef | 0.156 |
|---|
| CR2 |   | 5 | 360 | 957 | 938 | -0.042 | -0.089 | 0.101 | 0.018 | 0.015 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| NEDD8 |   | 4 | 440 | 1650 | 1612 | 0.250 | -0.085 | 0.251 | 0.211 | -0.073 |
|---|
| MAGEA10 |   | 1 | 1195 | 1879 | 1889 | undef | 0.022 | 0.152 | -0.011 | undef |
|---|
| ECHDC2 |   | 2 | 743 | 842 | 854 | -0.315 | 0.242 | -0.069 | 0.051 | 0.090 |
|---|
| FYN |   | 10 | 167 | 1 | 40 | -0.171 | -0.167 | 0.073 | -0.137 | 0.110 |
|---|
| GTPBP6 |   | 6 | 301 | 928 | 907 | -0.143 | -0.131 | -0.167 | 0.155 | -0.129 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| MAGEC1 |   | 2 | 743 | 842 | 854 | undef | 0.029 | 0.168 | 0.225 | -0.132 |
|---|
| SMURF1 |   | 5 | 360 | 682 | 671 | -0.222 | 0.236 | -0.049 | undef | -0.006 |
|---|
| STARD13 |   | 2 | 743 | 1879 | 1866 | -0.083 | 0.006 | 0.126 | 0.290 | 0.028 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| ANAPC5 |   | 1 | 1195 | 1879 | 1889 | -0.190 | 0.206 | -0.074 | -0.092 | 0.154 |
|---|
| USP4 |   | 7 | 256 | 1 | 47 | -0.093 | 0.147 | 0.097 | -0.392 | 0.054 |
|---|
| NSMCE4A |   | 2 | 743 | 842 | 854 | 0.177 | -0.116 | 0.290 | -0.022 | -0.091 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| SMAD7 |   | 5 | 360 | 1034 | 1004 | -0.035 | 0.131 | 0.169 | 0.061 | 0.147 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| CTAG1B |   | 1 | 1195 | 1879 | 1889 | 0.052 | 0.144 | 0.352 | -0.212 | -0.052 |
|---|
| SIPA1L3 |   | 3 | 557 | 648 | 647 | -0.016 | 0.039 | -0.062 | 0.136 | 0.049 |
|---|
| CUL5 |   | 3 | 557 | 1735 | 1718 | 0.051 | 0.187 | 0.221 | 0.017 | 0.216 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| RRAS2 |   | 7 | 256 | 1681 | 1633 | 0.155 | 0.076 | 0.289 | 0.044 | 0.031 |
|---|
| SMAD6 |   | 2 | 743 | 1879 | 1866 | -0.241 | 0.053 | 0.266 | -0.043 | 0.070 |
|---|
| ROCK2 |   | 1 | 1195 | 1879 | 1889 | -0.178 | 0.018 | 0.278 | 0.033 | 0.095 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PXN |   | 12 | 140 | 1 | 36 | -0.088 | 0.284 | -0.022 | 0.060 | 0.114 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| BCAR1 |   | 6 | 301 | 1 | 52 | -0.012 | 0.229 | undef | 0.223 | undef |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| SOX13 |   | 4 | 440 | 1034 | 1015 | -0.113 | 0.095 | -0.002 | 0.271 | -0.184 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| MAGED2 |   | 1 | 1195 | 1879 | 1889 | -0.104 | 0.198 | 0.022 | undef | -0.020 |
|---|
| TPR |   | 11 | 148 | 727 | 701 | -0.130 | 0.120 | -0.036 | -0.085 | 0.156 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GADD45A |   | 11 | 148 | 1351 | 1308 | -0.057 | 0.076 | -0.099 | 0.109 | 0.093 |
|---|
| FRAT1 |   | 1 | 1195 | 1879 | 1889 | -0.013 | -0.125 | 0.156 | undef | 0.078 |
|---|
| ACLY |   | 6 | 301 | 1260 | 1224 | -0.236 | 0.071 | 0.009 | 0.158 | 0.115 |
|---|
| RWDD2B |   | 1 | 1195 | 1879 | 1889 | -0.122 | 0.072 | 0.237 | -0.003 | 0.018 |
|---|
| IFI16 |   | 2 | 743 | 1681 | 1665 | 0.134 | -0.026 | 0.110 | -0.037 | -0.051 |
|---|
| ANAPC4 |   | 3 | 557 | 1083 | 1073 | 0.071 | 0.091 | undef | 0.030 | undef |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6473 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.263E-13 | 1.774E-09 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.31E-08 | 2.821E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.919E-08 | 3.192E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.338E-08 | 2.649E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 6.409E-08 | 3.132E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.094E-07 | 4.453E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.649E-07 | 5.755E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.03E-07 | 9.252E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 3.366E-07 | 9.137E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 4.645E-07 | 1.135E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 6.684E-07 | 1.485E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.803E-12 | 4.339E-09 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.965E-08 | 4.77E-05 |
|---|
| interphase | GO:0051325 |  | 5.065E-08 | 4.062E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 6.391E-08 | 3.844E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 7.093E-08 | 3.413E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.059E-07 | 4.247E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.82E-07 | 6.257E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.244E-07 | 6.749E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.514E-07 | 1.474E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 5.514E-07 | 1.327E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 7.006E-07 | 1.532E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.529E-12 | 1.042E-08 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.006E-07 | 1.157E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.138E-07 | 8.724E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.624E-07 | 9.339E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.113E-06 | 5.118E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.743E-06 | 6.681E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.811E-06 | 5.951E-04 |
|---|
| interphase | GO:0051325 |  | 2.037E-06 | 5.856E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.423E-06 | 1.386E-03 |
|---|
| ER overload response | GO:0006983 |  | 5.758E-06 | 1.324E-03 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 8.498E-06 | 1.777E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.389E-07 | 2.56E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.291E-07 | 2.111E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.869E-07 | 4.834E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.187E-06 | 5.47E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.424E-06 | 5.25E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.691E-06 | 8.267E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 5.941E-06 | 1.564E-03 |
|---|
| ER overload response | GO:0006983 |  | 6.283E-06 | 1.447E-03 |
|---|
| regulation of cell migration | GO:0030334 |  | 8.499E-06 | 1.74E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.144E-05 | 2.109E-03 |
|---|
| regulation of locomotion | GO:0040012 |  | 1.582E-05 | 2.65E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.529E-12 | 1.042E-08 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.006E-07 | 1.157E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.138E-07 | 8.724E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.624E-07 | 9.339E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.113E-06 | 5.118E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.743E-06 | 6.681E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.811E-06 | 5.951E-04 |
|---|
| interphase | GO:0051325 |  | 2.037E-06 | 5.856E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.423E-06 | 1.386E-03 |
|---|
| ER overload response | GO:0006983 |  | 5.758E-06 | 1.324E-03 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 8.498E-06 | 1.777E-03 |
|---|



