Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6467
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7556 | 3.407e-03 | 5.490e-04 | 7.661e-01 | 1.433e-06 |
|---|
| Loi | 0.2298 | 8.100e-02 | 1.155e-02 | 4.300e-01 | 4.024e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.581e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.473e-03 | 0.000e+00 |
|---|
| Wang | 0.2611 | 2.746e-03 | 4.444e-02 | 3.992e-01 | 4.872e-05 |
|---|
Expression data for subnetwork 6467 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
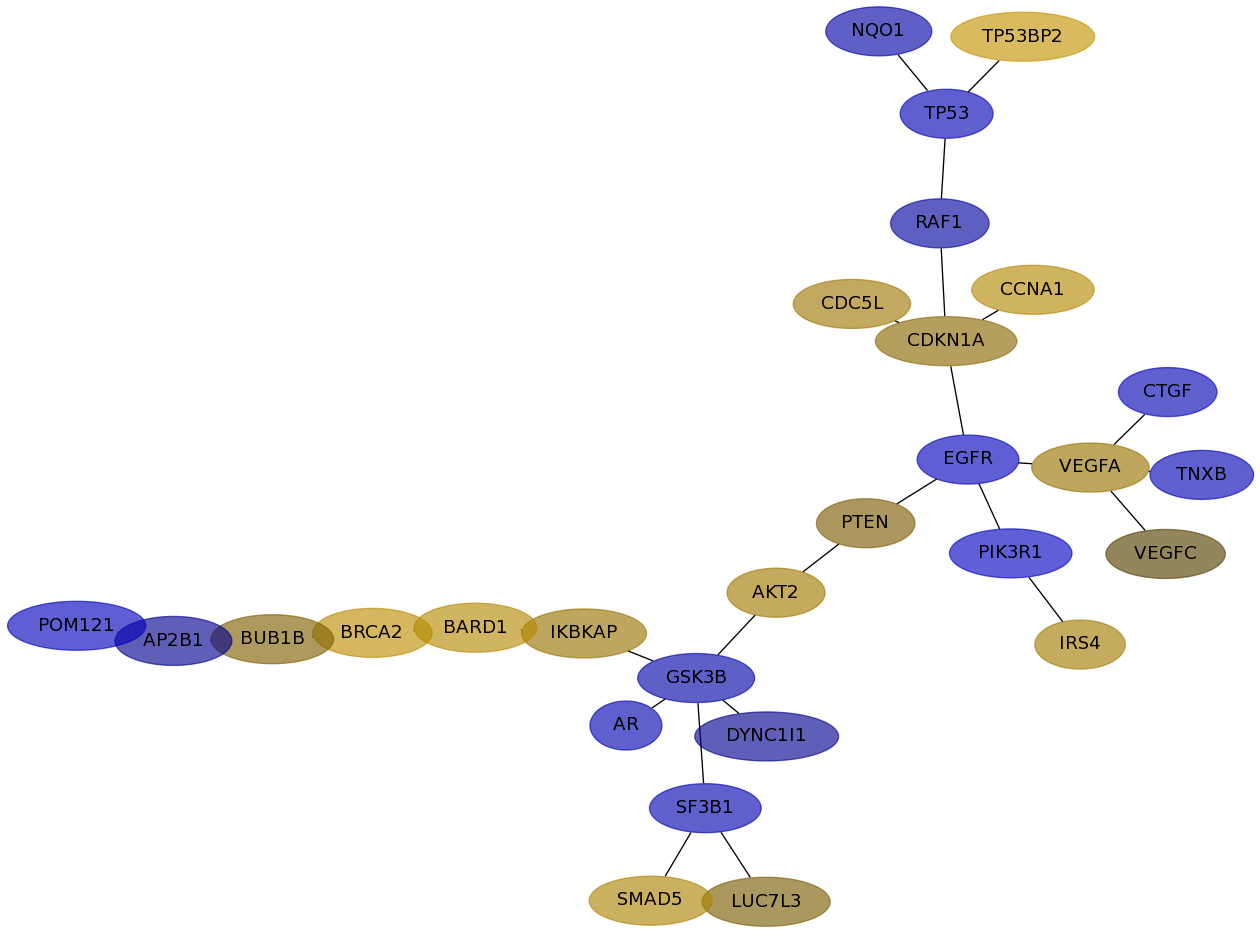
Score for each gene in subnetwork 6467 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| VEGFC |   | 2 | 743 | 1558 | 1545 | 0.018 | 0.123 | -0.010 | 0.271 | 0.245 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| RAF1 |   | 1 | 1195 | 1871 | 1881 | -0.105 | -0.004 | 0.033 | -0.086 | -0.227 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| TNXB |   | 4 | 440 | 727 | 723 | -0.169 | -0.076 | 0.160 | 0.182 | 0.173 |
|---|
| BUB1B |   | 4 | 440 | 1010 | 991 | 0.050 | -0.164 | 0.037 | -0.101 | 0.148 |
|---|
| SMAD5 |   | 2 | 743 | 1871 | 1852 | 0.144 | 0.008 | 0.039 | -0.012 | 0.039 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| IKBKAP |   | 10 | 167 | 412 | 408 | 0.089 | 0.184 | 0.142 | 0.105 | -0.017 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| DYNC1I1 |   | 5 | 360 | 614 | 612 | -0.069 | 0.013 | -0.086 | -0.045 | 0.088 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| POM121 |   | 7 | 256 | 780 | 763 | -0.197 | 0.101 | 0.079 | undef | 0.017 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| AP2B1 |   | 6 | 301 | 780 | 764 | -0.070 | -0.106 | 0.070 | 0.252 | 0.094 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| CDC5L |   | 2 | 743 | 1010 | 1007 | 0.102 | -0.114 | 0.107 | 0.024 | 0.133 |
|---|
GO Enrichment output for subnetwork 6467 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 2.531E-20 | 6.183E-17 |
|---|
| collagen metabolic process | GO:0032963 |  | 5.68E-15 | 6.938E-12 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 1.264E-14 | 1.029E-11 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 5.074E-14 | 3.099E-11 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 4.009E-13 | 1.959E-10 |
|---|
| aging | GO:0007568 |  | 9.863E-13 | 4.016E-10 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.605E-12 | 9.093E-10 |
|---|
| extracellular matrix organization | GO:0030198 |  | 3.734E-12 | 1.14E-09 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.313E-10 | 3.564E-08 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.496E-10 | 8.54E-08 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.932E-10 | 8.732E-08 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 1.448E-17 | 3.484E-14 |
|---|
| collagen metabolic process | GO:0032963 |  | 1.013E-12 | 1.219E-09 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 2.061E-12 | 1.653E-09 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 7.06E-12 | 4.247E-09 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.676E-10 | 8.063E-08 |
|---|
| extracellular matrix organization | GO:0030198 |  | 2.804E-10 | 1.124E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 4.72E-10 | 1.622E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 5.017E-10 | 1.509E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.168E-09 | 3.124E-07 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.333E-09 | 5.612E-07 |
|---|
| aging | GO:0007568 |  | 4.715E-09 | 1.031E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 1.398E-16 | 3.216E-13 |
|---|
| collagen metabolic process | GO:0032963 |  | 7.479E-12 | 8.6E-09 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 1.557E-11 | 1.194E-08 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 5.517E-11 | 3.172E-08 |
|---|
| extracellular matrix organization | GO:0030198 |  | 1.476E-09 | 6.789E-07 |
|---|
| aging | GO:0007568 |  | 2.277E-08 | 8.73E-06 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 2.5E-08 | 8.214E-06 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.592E-08 | 7.452E-06 |
|---|
| extracellular structure organization | GO:0043062 |  | 4.552E-08 | 1.163E-05 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 1.036E-07 | 2.382E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.825E-07 | 7.997E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ER overload response | GO:0006983 |  | 4.179E-07 | 7.702E-04 |
|---|
| angiogenesis | GO:0001525 |  | 8.276E-07 | 7.626E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.746E-06 | 1.072E-03 |
|---|
| blood vessel morphogenesis | GO:0048514 |  | 3.044E-06 | 1.402E-03 |
|---|
| negative regulation of cell cycle | GO:0045786 |  | 4.804E-06 | 1.771E-03 |
|---|
| blood vessel development | GO:0001568 |  | 5.64E-06 | 1.732E-03 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 5.899E-06 | 1.553E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.899E-06 | 1.359E-03 |
|---|
| vasculature development | GO:0001944 |  | 6.257E-06 | 1.281E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 9.35E-06 | 1.723E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 1.392E-05 | 2.333E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 1.398E-16 | 3.216E-13 |
|---|
| collagen metabolic process | GO:0032963 |  | 7.479E-12 | 8.6E-09 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 1.557E-11 | 1.194E-08 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 5.517E-11 | 3.172E-08 |
|---|
| extracellular matrix organization | GO:0030198 |  | 1.476E-09 | 6.789E-07 |
|---|
| aging | GO:0007568 |  | 2.277E-08 | 8.73E-06 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 2.5E-08 | 8.214E-06 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.592E-08 | 7.452E-06 |
|---|
| extracellular structure organization | GO:0043062 |  | 4.552E-08 | 1.163E-05 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 1.036E-07 | 2.382E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.825E-07 | 7.997E-05 |
|---|



