Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6461
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7557 | 3.400e-03 | 5.480e-04 | 7.658e-01 | 1.427e-06 |
|---|
| Loi | 0.2299 | 8.094e-02 | 1.154e-02 | 4.298e-01 | 4.014e-04 |
|---|
| Schmidt | 0.6725 | 0.000e+00 | 0.000e+00 | 4.588e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.477e-03 | 0.000e+00 |
|---|
| Wang | 0.2610 | 2.750e-03 | 4.447e-02 | 3.994e-01 | 4.884e-05 |
|---|
Expression data for subnetwork 6461 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
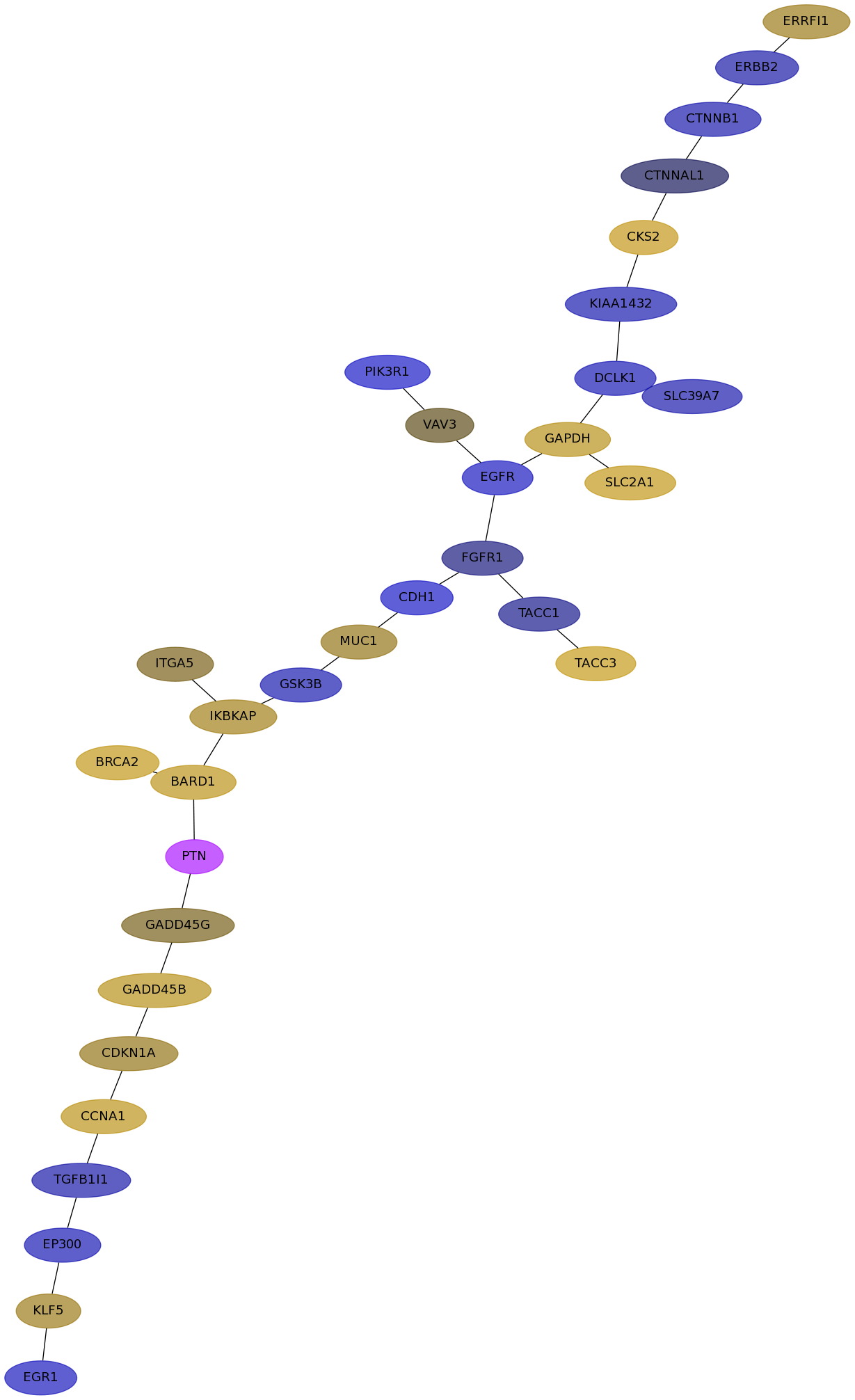
Score for each gene in subnetwork 6461 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| SLC2A1 |   | 7 | 256 | 236 | 237 | 0.205 | 0.143 | -0.050 | -0.023 | 0.086 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| IKBKAP |   | 10 | 167 | 412 | 408 | 0.089 | 0.184 | 0.142 | 0.105 | -0.017 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| KIAA1432 |   | 3 | 557 | 1681 | 1657 | -0.132 | -0.054 | undef | 0.108 | undef |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| SLC39A7 |   | 2 | 743 | 1845 | 1831 | -0.119 | 0.157 | -0.147 | 0.185 | 0.175 |
|---|
| ERRFI1 |   | 10 | 167 | 412 | 408 | 0.076 | 0.215 | undef | -0.043 | undef |
|---|
| TACC3 |   | 4 | 440 | 1077 | 1065 | 0.232 | -0.144 | 0.014 | 0.046 | 0.004 |
|---|
| CKS2 |   | 9 | 196 | 928 | 903 | 0.214 | -0.220 | 0.089 | -0.003 | -0.056 |
|---|
| VAV3 |   | 9 | 196 | 638 | 622 | 0.015 | 0.071 | -0.245 | 0.148 | 0.038 |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| TACC1 |   | 5 | 360 | 1077 | 1057 | -0.051 | 0.020 | -0.022 | 0.187 | 0.126 |
|---|
| CTNNAL1 |   | 4 | 440 | 682 | 678 | -0.014 | 0.073 | -0.008 | -0.131 | 0.004 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| DCLK1 |   | 7 | 256 | 141 | 151 | -0.145 | -0.006 | -0.048 | 0.188 | -0.035 |
|---|
| GADD45B |   | 8 | 222 | 318 | 325 | 0.158 | 0.122 | 0.067 | 0.060 | 0.212 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
GO Enrichment output for subnetwork 6461 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 7.756E-09 | 1.895E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.054E-08 | 4.952E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 6.487E-08 | 5.283E-05 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.944E-07 | 1.187E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 2.185E-07 | 1.068E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.818E-07 | 1.554E-04 |
|---|
| male meiosis | GO:0007140 |  | 6.098E-07 | 2.128E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.098E-07 | 1.862E-04 |
|---|
| dendrite morphogenesis | GO:0048813 |  | 6.098E-07 | 1.655E-04 |
|---|
| axon extension | GO:0048675 |  | 9.133E-07 | 2.231E-04 |
|---|
| central nervous system neuron axonogenesis | GO:0021955 |  | 1.303E-06 | 2.893E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 7.364E-09 | 1.772E-05 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 6.621E-08 | 7.965E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.059E-07 | 8.49E-05 |
|---|
| male meiosis | GO:0007140 |  | 2.951E-07 | 1.775E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 2.951E-07 | 1.42E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 3.166E-07 | 1.269E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.154E-07 | 1.772E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 8.232E-07 | 2.476E-04 |
|---|
| dendrite morphogenesis | GO:0048813 |  | 8.232E-07 | 2.201E-04 |
|---|
| axon extension | GO:0048675 |  | 1.233E-06 | 2.966E-04 |
|---|
| central nervous system neuron axonogenesis | GO:0021955 |  | 1.758E-06 | 3.845E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.73E-08 | 1.778E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.992E-07 | 2.291E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 4.392E-07 | 3.367E-04 |
|---|
| male meiosis | GO:0007140 |  | 5.854E-07 | 3.366E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.854E-07 | 2.693E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 5.854E-07 | 2.244E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.632E-06 | 5.362E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.632E-06 | 4.691E-04 |
|---|
| regulation of protein transport | GO:0051223 |  | 3.99E-06 | 1.02E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 6.645E-06 | 1.528E-03 |
|---|
| regulation of protein localization | GO:0032880 |  | 6.933E-06 | 1.45E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.589E-08 | 4.772E-05 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 3.914E-08 | 3.607E-05 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 8.154E-08 | 5.009E-05 |
|---|
| regulation of protein transport | GO:0051223 |  | 1.302E-06 | 5.999E-04 |
|---|
| regulation of protein localization | GO:0032880 |  | 1.88E-06 | 6.931E-04 |
|---|
| negative regulation of nucleocytoplasmic transport | GO:0046823 |  | 5.651E-06 | 1.736E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 7.75E-06 | 2.04E-03 |
|---|
| activation of MAPKK activity | GO:0000186 |  | 7.75E-06 | 1.785E-03 |
|---|
| negative regulation of intracellular transport | GO:0032387 |  | 1.337E-05 | 2.737E-03 |
|---|
| regulation of cellular localization | GO:0060341 |  | 1.371E-05 | 2.527E-03 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 1.452E-05 | 2.433E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.73E-08 | 1.778E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.992E-07 | 2.291E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 4.392E-07 | 3.367E-04 |
|---|
| male meiosis | GO:0007140 |  | 5.854E-07 | 3.366E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.854E-07 | 2.693E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 5.854E-07 | 2.244E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.632E-06 | 5.362E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.632E-06 | 4.691E-04 |
|---|
| regulation of protein transport | GO:0051223 |  | 3.99E-06 | 1.02E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 6.645E-06 | 1.528E-03 |
|---|
| regulation of protein localization | GO:0032880 |  | 6.933E-06 | 1.45E-03 |
|---|



