Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6460
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7558 | 3.397e-03 | 5.470e-04 | 7.657e-01 | 1.423e-06 |
|---|
| Loi | 0.2298 | 8.102e-02 | 1.156e-02 | 4.300e-01 | 4.027e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.581e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.479e-03 | 0.000e+00 |
|---|
| Wang | 0.2611 | 2.749e-03 | 4.446e-02 | 3.993e-01 | 4.881e-05 |
|---|
Expression data for subnetwork 6460 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
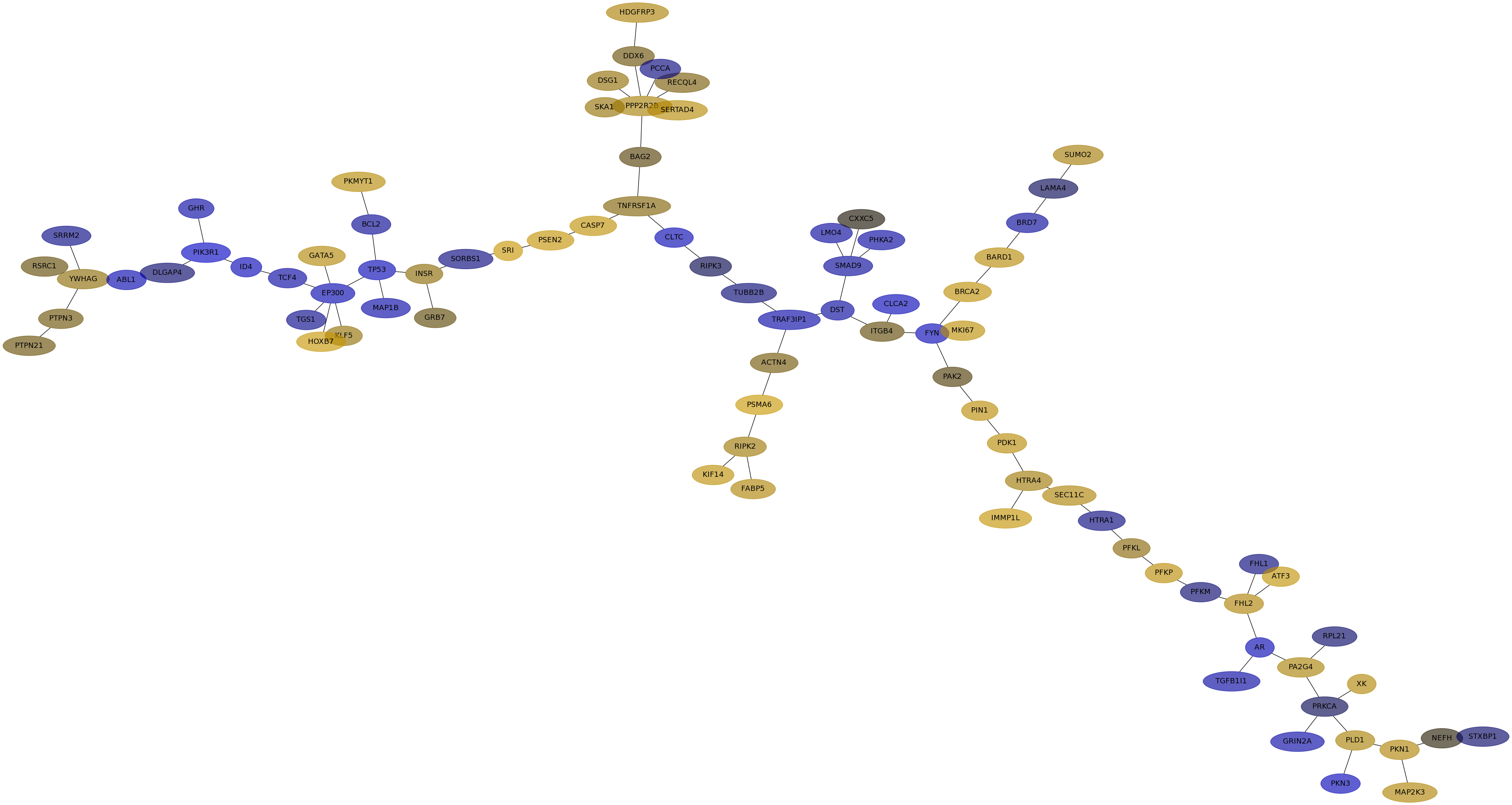
Score for each gene in subnetwork 6460 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| HOXB7 |   | 6 | 301 | 1 | 52 | 0.267 | -0.139 | -0.083 | 0.085 | -0.051 |
|---|
| HTRA1 |   | 7 | 256 | 83 | 102 | -0.046 | 0.184 | 0.051 | 0.132 | 0.224 |
|---|
| PTPN21 |   | 5 | 360 | 1717 | 1677 | 0.027 | 0.177 | 0.214 | 0.205 | 0.119 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| HDGFRP3 |   | 1 | 1195 | 1830 | 1841 | 0.136 | -0.040 | 0.160 | 0.126 | 0.143 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| MAP2K3 |   | 1 | 1195 | 1830 | 1841 | 0.150 | 0.041 | 0.131 | undef | -0.045 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| PA2G4 |   | 1 | 1195 | 1830 | 1841 | 0.130 | -0.066 | 0.023 | 0.001 | -0.022 |
|---|
| PSMA6 |   | 5 | 360 | 842 | 828 | 0.280 | -0.118 | -0.018 | undef | -0.091 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| LMO4 |   | 1 | 1195 | 1830 | 1841 | -0.094 | 0.071 | 0.017 | -0.045 | -0.123 |
|---|
| KIF14 |   | 1 | 1195 | 1830 | 1841 | 0.205 | -0.056 | 0.041 | undef | 0.010 |
|---|
| SRI |   | 7 | 256 | 614 | 601 | 0.270 | -0.040 | 0.181 | 0.185 | 0.057 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| PHKA2 |   | 3 | 557 | 991 | 982 | -0.107 | 0.059 | -0.153 | 0.250 | 0.041 |
|---|
| SRRM2 |   | 2 | 743 | 1396 | 1391 | -0.054 | 0.101 | -0.080 | 0.024 | 0.111 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| CXXC5 |   | 1 | 1195 | 1830 | 1841 | 0.003 | 0.137 | undef | 0.090 | undef |
|---|
| PDK1 |   | 10 | 167 | 1 | 40 | 0.183 | -0.138 | -0.041 | -0.265 | -0.126 |
|---|
| RIPK2 |   | 2 | 743 | 141 | 197 | 0.101 | -0.133 | -0.032 | -0.063 | 0.034 |
|---|
| SMAD9 |   | 9 | 196 | 296 | 285 | -0.090 | -0.153 | 0.055 | 0.081 | -0.059 |
|---|
| SERTAD4 |   | 8 | 222 | 1268 | 1225 | 0.173 | 0.169 | undef | -0.050 | undef |
|---|
| MKI67 |   | 5 | 360 | 1 | 56 | 0.205 | -0.123 | 0.176 | -0.086 | 0.018 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| FABP5 |   | 3 | 557 | 1534 | 1516 | 0.152 | -0.177 | 0.225 | -0.118 | 0.141 |
|---|
| PKMYT1 |   | 7 | 256 | 1510 | 1463 | 0.175 | -0.143 | 0.046 | 0.105 | -0.070 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| FYN |   | 10 | 167 | 1 | 40 | -0.171 | -0.167 | 0.073 | -0.137 | 0.110 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| CLCA2 |   | 9 | 196 | 412 | 411 | -0.176 | 0.243 | -0.153 | 0.191 | -0.000 |
|---|
| CLTC |   | 3 | 557 | 983 | 979 | -0.155 | 0.072 | -0.095 | 0.179 | 0.048 |
|---|
| TRAF3IP1 |   | 9 | 196 | 83 | 99 | -0.113 | -0.063 | -0.088 | 0.276 | undef |
|---|
| HTRA4 |   | 8 | 222 | 83 | 101 | 0.102 | undef | undef | undef | undef |
|---|
| TNFRSF1A |   | 8 | 222 | 412 | 413 | 0.050 | 0.094 | -0.014 | 0.214 | -0.045 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| DSG1 |   | 10 | 167 | 658 | 636 | 0.072 | -0.075 | 0.252 | -0.061 | 0.068 |
|---|
| IMMP1L |   | 2 | 743 | 1830 | 1815 | 0.243 | -0.028 | undef | undef | undef |
|---|
| TGS1 |   | 5 | 360 | 780 | 767 | -0.060 | -0.044 | 0.232 | undef | 0.269 |
|---|
| GATA5 |   | 8 | 222 | 366 | 373 | 0.161 | -0.031 | undef | undef | undef |
|---|
| PFKP |   | 6 | 301 | 83 | 105 | 0.195 | -0.076 | -0.040 | undef | 0.094 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| DLGAP4 |   | 7 | 256 | 727 | 714 | -0.029 | 0.180 | -0.201 | 0.135 | -0.043 |
|---|
| PTPN3 |   | 11 | 148 | 658 | 635 | 0.029 | 0.060 | -0.067 | 0.225 | -0.034 |
|---|
| PKN1 |   | 5 | 360 | 1674 | 1634 | 0.150 | -0.143 | -0.011 | 0.084 | -0.002 |
|---|
| BAG2 |   | 4 | 440 | 1793 | 1755 | 0.016 | 0.178 | 0.152 | -0.249 | 0.083 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| RECQL4 |   | 2 | 743 | 1830 | 1815 | 0.039 | -0.211 | 0.069 | -0.095 | -0.053 |
|---|
| DDX6 |   | 4 | 440 | 1688 | 1656 | 0.028 | 0.162 | 0.157 | -0.035 | 0.084 |
|---|
| TCF4 |   | 2 | 743 | 727 | 734 | -0.109 | 0.229 | 0.063 | 0.025 | 0.097 |
|---|
| PFKL |   | 7 | 256 | 83 | 102 | 0.056 | 0.106 | -0.173 | 0.160 | -0.158 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| GRIN2A |   | 12 | 140 | 179 | 192 | -0.113 | -0.068 | 0.162 | 0.125 | undef |
|---|
| STXBP1 |   | 5 | 360 | 1629 | 1587 | -0.027 | 0.078 | 0.180 | 0.143 | 0.043 |
|---|
| PCCA |   | 2 | 743 | 1825 | 1808 | -0.045 | 0.042 | 0.100 | 0.145 | -0.016 |
|---|
| RPL21 |   | 1 | 1195 | 1830 | 1841 | -0.026 | 0.106 | 0.270 | -0.222 | -0.130 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SEC11C |   | 2 | 743 | 806 | 806 | 0.141 | -0.215 | undef | undef | undef |
|---|
| NEFH |   | 1 | 1195 | 1830 | 1841 | 0.005 | 0.107 | -0.079 | -0.003 | -0.078 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| SKA1 |   | 2 | 743 | 1358 | 1360 | 0.081 | -0.085 | 0.037 | undef | -0.070 |
|---|
| PFKM |   | 6 | 301 | 83 | 105 | -0.030 | -0.079 | 0.293 | 0.023 | 0.118 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CASP7 |   | 6 | 301 | 614 | 602 | 0.216 | 0.095 | 0.255 | undef | 0.035 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
GO Enrichment output for subnetwork 6460 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.661E-07 | 6.501E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 1.132E-06 | 1.382E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.015E-06 | 1.641E-03 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 6.549E-06 | 4E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 6.993E-06 | 3.417E-03 |
|---|
| B cell lineage commitment | GO:0002326 |  | 6.993E-06 | 2.847E-03 |
|---|
| T cell lineage commitment | GO:0002360 |  | 6.993E-06 | 2.44E-03 |
|---|
| cell aging | GO:0007569 |  | 7.968E-06 | 2.433E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.359E-05 | 3.689E-03 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.599E-05 | 3.905E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.664E-05 | 3.695E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.112E-07 | 9.893E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 1.524E-06 | 1.834E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.527E-06 | 2.027E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 8.761E-06 | 5.27E-03 |
|---|
| B cell lineage commitment | GO:0002326 |  | 8.761E-06 | 4.216E-03 |
|---|
| T cell lineage commitment | GO:0002360 |  | 8.761E-06 | 3.513E-03 |
|---|
| cell aging | GO:0007569 |  | 8.801E-06 | 3.025E-03 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 8.801E-06 | 2.647E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.823E-05 | 4.875E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.083E-05 | 5.012E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.083E-05 | 4.556E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 8.929E-07 | 2.054E-03 |
|---|
| response to gamma radiation | GO:0010332 |  | 2.134E-06 | 2.455E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 5.522E-06 | 4.233E-03 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.22E-05 | 7.015E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.909E-05 | 8.782E-03 |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.909E-05 | 7.318E-03 |
|---|
| T cell lineage commitment | GO:0002360 |  | 1.909E-05 | 6.273E-03 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 1.984E-05 | 5.704E-03 |
|---|
| cell aging | GO:0007569 |  | 2.435E-05 | 6.223E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.036E-05 | 6.983E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 3.036E-05 | 6.348E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| T cell lineage commitment | GO:0002360 |  | 4.781E-06 | 8.811E-03 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 6.423E-06 | 5.918E-03 |
|---|
| cell junction assembly | GO:0034329 |  | 8.055E-06 | 4.948E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 9.508E-06 | 4.381E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 9.508E-06 | 3.505E-03 |
|---|
| response to gamma radiation | GO:0010332 |  | 1.654E-05 | 5.082E-03 |
|---|
| cell junction organization | GO:0034330 |  | 2.403E-05 | 6.327E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.632E-05 | 6.064E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.632E-05 | 5.39E-03 |
|---|
| positive regulation of growth | GO:0045927 |  | 2.892E-05 | 5.33E-03 |
|---|
| regulation of catabolic process | GO:0009894 |  | 3.224E-05 | 5.401E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 8.929E-07 | 2.054E-03 |
|---|
| response to gamma radiation | GO:0010332 |  | 2.134E-06 | 2.455E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 5.522E-06 | 4.233E-03 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.22E-05 | 7.015E-03 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.909E-05 | 8.782E-03 |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.909E-05 | 7.318E-03 |
|---|
| T cell lineage commitment | GO:0002360 |  | 1.909E-05 | 6.273E-03 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 1.984E-05 | 5.704E-03 |
|---|
| cell aging | GO:0007569 |  | 2.435E-05 | 6.223E-03 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.036E-05 | 6.983E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 3.036E-05 | 6.348E-03 |
|---|



